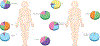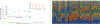Current understanding of the human microbiome - PubMed (original) (raw)
Review
Current understanding of the human microbiome
Jack A Gilbert et al. Nat Med. 2018.
Abstract
Our understanding of the link between the human microbiome and disease, including obesity, inflammatory bowel disease, arthritis and autism, is rapidly expanding. Improvements in the throughput and accuracy of DNA sequencing of the genomes of microbial communities that are associated with human samples, complemented by analysis of transcriptomes, proteomes, metabolomes and immunomes and by mechanistic experiments in model systems, have vastly improved our ability to understand the structure and function of the microbiome in both diseased and healthy states. However, many challenges remain. In this review, we focus on studies in humans to describe these challenges and propose strategies that leverage existing knowledge to move rapidly from correlation to causation and ultimately to translation into therapies.
Figures
Figure 1:
The human microbiome is highly personalized. Understanding the relevance of the differing microbiota between individuals is confounded by the uniqueness of an individuals’ microbiome. The different colours in the pie charts represent different species.
Figure 2:
The dynamics of the human microbiome. The human microbiome has been shown to be highly dynamic. A) Taking a “representative” sample of a human microbiome at any given site is challenging because while the microbiome is known to settle after birth (green line), the composition can vary both over short term and long term timescales (orange line and blue line respectively). B) The effect of the rate of change of the varying species on the ability to take a representative sample as indicated by the black line is shown.
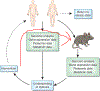
Figure 3.
Towards further understanding and developing therapies from microbiome data. The iterative cycle of analysis, interpretation and translational intervention that facilitate moving microbiome research out of correlative observation and into therapeutic treatments is shown.
Similar articles
- Human Microbiome and Learning Healthcare Systems: Integrating Research and Precision Medicine for Inflammatory Bowel Disease.
Chuong KH, Mack DR, Stintzi A, O'Doherty KC. Chuong KH, et al. OMICS. 2018 Feb;22(2):119-126. doi: 10.1089/omi.2016.0185. Epub 2017 Mar 10. OMICS. 2018. PMID: 28282257 Free PMC article. Review. - Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer.
Rajpoot M, Sharma AK, Sharma A, Gupta GK. Rajpoot M, et al. Semin Cancer Biol. 2018 Oct;52(Pt 1):1-8. doi: 10.1016/j.semcancer.2018.02.003. Epub 2018 Feb 6. Semin Cancer Biol. 2018. PMID: 29425888 Review. - De novo Assembly Vastly Expands the Known Microbial Universe.
Minot SS. Minot SS. Trends Microbiol. 2019 May;27(5):385-386. doi: 10.1016/j.tim.2019.02.007. Epub 2019 Mar 11. Trends Microbiol. 2019. PMID: 30871856 - The Microbiome and Its Potential for Pharmacology.
Chavira A, Belda-Ferre P, Kosciolek T, Ali F, Dorrestein PC, Knight R. Chavira A, et al. Handb Exp Pharmacol. 2019;260:301-326. doi: 10.1007/164_2019_317. Handb Exp Pharmacol. 2019. PMID: 31820171 Review. - Systems biology of the human microbiome.
Peñalver Bernabé B, Cralle L, Gilbert JA. Peñalver Bernabé B, et al. Curr Opin Biotechnol. 2018 Jun;51:146-153. doi: 10.1016/j.copbio.2018.01.018. Epub 2018 Feb 14. Curr Opin Biotechnol. 2018. PMID: 29453029 Review.
Cited by
- Validation and standardization of DNA extraction and library construction methods for metagenomics-based human fecal microbiome measurements.
Tourlousse DM, Narita K, Miura T, Sakamoto M, Ohashi A, Shiina K, Matsuda M, Miura D, Shimamura M, Ohyama Y, Yamazoe A, Uchino Y, Kameyama K, Arioka S, Kataoka J, Hisada T, Fujii K, Takahashi S, Kuroiwa M, Rokushima M, Nishiyama M, Tanaka Y, Fuchikami T, Aoki H, Kira S, Koyanagi R, Naito T, Nishiwaki M, Kumagai H, Konda M, Kasahara K, Ohkuma M, Kawasaki H, Sekiguchi Y, Terauchi J. Tourlousse DM, et al. Microbiome. 2021 Apr 29;9(1):95. doi: 10.1186/s40168-021-01048-3. Microbiome. 2021. PMID: 33910647 Free PMC article. - The Oral and Gut Bacterial Microbiomes: Similarities, Differences, and Connections.
Maki KA, Kazmi N, Barb JJ, Ames N. Maki KA, et al. Biol Res Nurs. 2021 Jan;23(1):7-20. doi: 10.1177/1099800420941606. Epub 2020 Jul 21. Biol Res Nurs. 2021. PMID: 32691605 Free PMC article. Review. - Exploring the microbiome: Uncovering the link with lung cancer and implications for diagnosis and treatment.
Yi J, Xiang J, Tang J. Yi J, et al. Chin Med J Pulm Crit Care Med. 2023 Sep 15;1(3):161-170. doi: 10.1016/j.pccm.2023.08.003. eCollection 2023 Sep. Chin Med J Pulm Crit Care Med. 2023. PMID: 39171127 Free PMC article. Review. - Specific gut microbiota may increase the risk of erectile dysfunction: a two-sample Mendelian randomization study.
Su Q, Long Y, Luo Y, Jiang T, Zheng L, Wang K, Tang Q. Su Q, et al. Front Endocrinol (Lausanne). 2023 Dec 18;14:1216746. doi: 10.3389/fendo.2023.1216746. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 38192423 Free PMC article. - Autologous Faecal Microbiota Transplantation to Improve Outcomes of Haematopoietic Stem Cell Transplantation: Results of a Single-Centre Feasibility Study.
Li A, Bowen JM, Ball IA, Wilson S, Yong A, Yeung DT, Lee CH, Bryant RV, Costello SP, Ryan FJ, Wardill HR. Li A, et al. Biomedicines. 2023 Dec 11;11(12):3274. doi: 10.3390/biomedicines11123274. Biomedicines. 2023. PMID: 38137495 Free PMC article.
References
- Sender R, Fuchs S. & Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164, 337–340 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources