Yes-associated protein (YAP) in pancreatic cancer: at the epicenter of a targetable signaling network associated with patient survival - PubMed (original) (raw)
Yes-associated protein (YAP) in pancreatic cancer: at the epicenter of a targetable signaling network associated with patient survival
Enrique Rozengurt et al. Signal Transduct Target Ther. 2018.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is generally a fatal disease with no efficacious treatment modalities. Elucidation of signaling mechanisms that will lead to the identification of novel targets for therapy and chemoprevention is urgently needed. Here, we review the role of Yes-associated protein (YAP) and WW-domain-containing Transcriptional co-Activator with a PDZ-binding motif (TAZ) in the development of PDAC. These oncogenic proteins are at the center of a signaling network that involves multiple upstream signals and downstream YAP-regulated genes. We also discuss the clinical significance of the YAP signaling network in PDAC using a recently published interactive open-access database (www.proteinatlas.org/pathology) that allows genome-wide exploration of the impact of individual proteins on survival outcomes. Multiple YAP/TEAD-regulated genes, including AJUBA, ANLN, AREG, ARHGAP29, AURKA, BUB1, CCND1, CDK6, CXCL5, EDN2, DKK1, FOSL1,FOXM1, HBEGF, IGFBP2, JAG1, NOTCH2, RHAMM, RRM2, SERP1, and ZWILCH, are associated with unfavorable survival of PDAC patients. Similarly, components of AP-1 that synergize with YAP (FOSL1), growth factors (TGFα, EPEG, and HBEGF), a specific integrin (ITGA2), heptahelical receptors (P2Y 2 R, GPR87) and an inhibitor of the Hippo pathway (MUC1), all of which stimulate YAP activity, are associated with unfavorable survival of PDAC patients. By contrast, YAP inhibitory pathways (STRAD/LKB-1/AMPK, PKA/LATS, and TSC/mTORC1) indicate a favorable prognosis. These associations emphasize that the YAP signaling network correlates with poor survival of pancreatic cancer patients. We conclude that the YAP pathway is a major determinant of clinical aggressiveness in PDAC patients and a target for therapeutic and preventive strategies in this disease.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
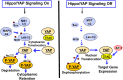
Fig. 1
Hippo signaling phosphorylates YAP and regulates its nuclear/cytoplasmic distribution. When Hippo signaling is active (e.g., in response to cell density, polarity signals, or mechanical cues) the Mst1/2 kinases, in complex with Sav1, phosphorylate and activate Lats1/2 in complex with its regulatory protein MOB1/2. In addition to Mst1/2, MAP4Ks act as alternative kinases that phosphorylate Lats1/2. In turn, Lats1/2 phosphorylates YAP on highly conserved residues (in red) located within a consensus sequence that is phosphorylated by Lats1/2 (HXRXXS). The phosphorylation of YAP at Ser-127 promotes its cytoplasmic retention, whereas phosphorylation at Ser-397 induces degradation. When the Hippo pathway is off, YAP is dephosphorylated and translocated into the nucleus where YAP binds and activates the TEAD transcription factors and stimulates the expression of multiple genes. Additional details are provided in the text
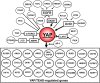
Fig. 2
YAP is at the epicenter of a signaling network. Growth factors (TGFα, EPEG, and HBEGF) induce EGFR signaling leading to KRAS activation, which in turn, stimulates YAP activation and increased expression. Other tyrosine kinase receptors, a specific integrin (ITGA2), multiple GPCRs, an inhibitor of Hippo pathway (MUC1) and actin polymerization via multiple pathways also stimulate YAP activity. Activation of YAP stimulates its coupling with TEAD, thereby promoting the expression of multiple YAP/TEAD-regulated genes that were identified in screens in different cell types. Reference to the study or studies connecting each gene to YAP/TAZ, as well as further details concerning the network are in the text
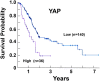
Fig. 3
Kaplan–Meier plots for YAP expression in PDAC. The image was reproduced from the Human Protein Atlas (version 17) available at
. The link is:
http://www.proteinatlas.org/ENSG00000137693YAP1/pathology/tissue/pancreatic±cancer.
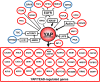
Fig. 4
YAP signaling is associated with unfavorable prognosis for PDAC. Multiple YAP/TEAD-regulated genes are associated with unfavorable survival of PDAC patients (indicated in red). Growth factors (TGFα, EPEG, and HBEGF), a specific integrin (ITGA2), GPCRs (P2Y 2 R, GPR87) or an inhibitor of the Hippo pathway (MUC1) that stimulate YAP activity are also associated with unfavorable survival in PDAC. Conversely, YAP inhibitory pathways, including STRAD/LKB-1, PKA/LATS, and TSC/mTORC1 are associated with a favorable prognosis (indicated in blue). The key feature is that each component of the network has an impact on survival of PDAC patients, as derived from the Pathology Atlas, as well as additional references cited in the text. An unfavorable prognosis for PDAC is in red and a favorable prognosis for PDAC is in blue. All prognostic associations are highly statistically significant (p < 0.001). Further details are in the text
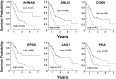
Fig. 5
Kaplan–Meier plots for gene expression of the YAP signaling network in PDAC. Images were reproduced from the Human Protein Atlas (version 17) available at
. The links to the specific genes shown are as follows: AHNAK,
http://www.proteinatlas.org/ENSG00000124942AHNAK/pathology/tissue/pancreatic±cancer
, ANLN,
http://www.proteinatlas.org/ENSG00000011426ANLN/pathology/tissue/pancreatic±cancer
CDK6,
http://www.proteinatlas.org/ENSG00000105810-CDK6/pathology/tissue/pancreatic±cancer
EPS8,
http://www.proteinatlas.org/ENSG00000151491-EPS8/pathology/tissue/pancreatic±cancer
JAG1,
http://www.proteinatlas.org/ENSG00000101384-JAG1/pathology/tissue/pancreatic±cancer
PKA,
http://www.proteinatlas.org/ENSG00000072062-PRKACA/pathology/tissue/pancreatic±cancer
Similar articles
- Central role of Yes-associated protein and WW-domain-containing transcriptional co-activator with PDZ-binding motif in pancreatic cancer development.
Rozengurt E, Eibl G. Rozengurt E, et al. World J Gastroenterol. 2019 Apr 21;25(15):1797-1816. doi: 10.3748/wjg.v25.i15.1797. World J Gastroenterol. 2019. PMID: 31057295 Free PMC article. Review. - Biological Significance of YAP/TAZ in Pancreatic Ductal Adenocarcinoma.
Hayashi H, Uemura N, Zhao L, Matsumura K, Sato H, Shiraishi Y, Baba H. Hayashi H, et al. Front Oncol. 2021 Jul 29;11:700315. doi: 10.3389/fonc.2021.700315. eCollection 2021. Front Oncol. 2021. PMID: 34395269 Free PMC article. Review. - YAP in pancreatic cancer: oncogenic role and therapeutic strategy.
Mao W, Mai J, Peng H, Wan J, Sun T. Mao W, et al. Theranostics. 2021 Jan 1;11(4):1753-1762. doi: 10.7150/thno.53438. eCollection 2021. Theranostics. 2021. PMID: 33408779 Free PMC article. Review. - Targeting the Hippo Signaling Pathway for Tissue Regeneration and Cancer Therapy.
Juan WC, Hong W. Juan WC, et al. Genes (Basel). 2016 Aug 30;7(9):55. doi: 10.3390/genes7090055. Genes (Basel). 2016. PMID: 27589805 Free PMC article. Review. - Yes-associated protein (YAP) and transcriptional coactivator with a PDZ-binding motif (TAZ): a nexus between hypoxia and cancer.
Zhao C, Zeng C, Ye S, Dai X, He Q, Yang B, Zhu H. Zhao C, et al. Acta Pharm Sin B. 2020 Jun;10(6):947-960. doi: 10.1016/j.apsb.2019.12.010. Epub 2019 Dec 19. Acta Pharm Sin B. 2020. PMID: 32642404 Free PMC article. Review.
Cited by
- Hyaluronic acid metabolism and chemotherapy resistance: recent advances and therapeutic potential.
Liu Z, Hou P, Fang J, Shao C, Shi Y, Melino G, Peschiaroli A. Liu Z, et al. Mol Oncol. 2024 Sep;18(9):2087-2106. doi: 10.1002/1878-0261.13551. Epub 2024 Mar 21. Mol Oncol. 2024. PMID: 37953485 Free PMC article. Review. - Sequestration of Intestinal Acidic Toxins by Cationic Resin Attenuates Pancreatic Cancer Progression through Promoting Autophagic Flux for YAP Degradation.
Zhao G, Zhang T, Liu W, Edderkaoui M, Hu R, Li J, Pandol SJ, Fu X, Han YP. Zhao G, et al. Cancers (Basel). 2022 Mar 10;14(6):1407. doi: 10.3390/cancers14061407. Cancers (Basel). 2022. PMID: 35326559 Free PMC article. - Cancer-Associated Fibroblasts in Gastrointestinal Cancers: Unveiling Their Dynamic Roles in the Tumor Microenvironment.
Al-Bzour NN, Al-Bzour AN, Ababneh OE, Al-Jezawi MM, Saeed A, Saeed A. Al-Bzour NN, et al. Int J Mol Sci. 2023 Nov 19;24(22):16505. doi: 10.3390/ijms242216505. Int J Mol Sci. 2023. PMID: 38003695 Free PMC article. Review. - Role of cell rearrangement and related signaling pathways in the dynamic process of tip cell selection.
Guo Y, Zhang S, Wang D, Heng BC, Deng X. Guo Y, et al. Cell Commun Signal. 2024 Jan 9;22(1):24. doi: 10.1186/s12964-023-01364-1. Cell Commun Signal. 2024. PMID: 38195565 Free PMC article. Review. - Abemaciclib Is Effective Against Pancreatic Cancer Cells and Synergizes with HuR and YAP1 Inhibition.
Dhir T, Schultz CW, Jain A, Brown SZ, Haber A, Goetz A, Xi C, Su GH, Xu L, Posey J 3rd, Jiang W, Yeo CJ, Golan T, Pishvaian MJ, Brody JR. Dhir T, et al. Mol Cancer Res. 2019 Oct;17(10):2029-2041. doi: 10.1158/1541-7786.MCR-19-0589. Epub 2019 Aug 5. Mol Cancer Res. 2019. PMID: 31383722 Free PMC article.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics. Cancer J. Clin. 2017;67:7–30. - PubMed
- Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous