Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7 - PubMed (original) (raw)
Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7
Andrew Rambaut et al. Syst Biol. 2018.
Abstract
Bayesian inference of phylogeny using Markov chain Monte Carlo (MCMC) plays a central role in understanding evolutionary history from molecular sequence data. Visualizing and analyzing the MCMC-generated samples from the posterior distribution is a key step in any non-trivial Bayesian inference. We present the software package Tracer (version 1.7) for visualizing and analyzing the MCMC trace files generated through Bayesian phylogenetic inference. Tracer provides kernel density estimation, multivariate visualization, demographic trajectory reconstruction, conditional posterior distribution summary, and more. Tracer is open-source and available at http://beast.community/tracer.
Figures
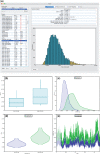
Figure 1.
Overview of Tracer functionality and individual parameter visualizations: a) Main Tracer panel upon loading a single trace file; b) boxplot representation of two continuous parameters; corresponding c) kernel density estimates; d) violin plots; e) the actual traces connecting the parameter values visited by the Markov chain.
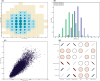
Figure 2.
Multi-parameter visualizations of: a) the joint probability distribution of two integer variables through a bubble chart; b) the marginal density of multiple integer or categorical variables through frequency plots; c) two continuous variables through a classic scatter or correlation plot; d) multiple () continuous variables using large correlation matrices.
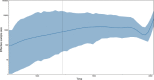
Figure 3.
Estimating the effective population sizes over time using a Bayesian skygrid demographic reconstruction for rabies virus in North America.
Similar articles
- AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics.
Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. Nylander JA, et al. Bioinformatics. 2008 Feb 15;24(4):581-3. doi: 10.1093/bioinformatics/btm388. Epub 2007 Aug 30. Bioinformatics. 2008. PMID: 17766271 - Bayesian phylogenetics with BEAUti and the BEAST 1.7.
Drummond AJ, Suchard MA, Xie D, Rambaut A. Drummond AJ, et al. Mol Biol Evol. 2012 Aug;29(8):1969-73. doi: 10.1093/molbev/mss075. Epub 2012 Feb 25. Mol Biol Evol. 2012. PMID: 22367748 Free PMC article. - RWTY (R We There Yet): An R Package for Examining Convergence of Bayesian Phylogenetic Analyses.
Warren DL, Geneva AJ, Lanfear R. Warren DL, et al. Mol Biol Evol. 2017 Apr 1;34(4):1016-1020. doi: 10.1093/molbev/msw279. Mol Biol Evol. 2017. PMID: 28087773 - A biologist's guide to Bayesian phylogenetic analysis.
Nascimento FF, Reis MD, Yang Z. Nascimento FF, et al. Nat Ecol Evol. 2017 Oct;1(10):1446-1454. doi: 10.1038/s41559-017-0280-x. Epub 2017 Sep 21. Nat Ecol Evol. 2017. PMID: 28983516 Free PMC article. Review. - Scalable Bayesian phylogenetics.
Fisher AA, Hassler GW, Ji X, Baele G, Suchard MA, Lemey P. Fisher AA, et al. Philos Trans R Soc Lond B Biol Sci. 2022 Oct 10;377(1861):20210242. doi: 10.1098/rstb.2021.0242. Epub 2022 Aug 22. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35989603 Free PMC article. Review.
Cited by
- Genetic diversity in a unique population of dugong (Dugong dugon) along the sea coasts of Thailand.
Poommouang A, Kriangwanich W, Buddhachat K, Brown JL, Piboon P, Chomdej S, Kampuansai J, Mekchay S, Kaewmong P, Kittiwattanawong K, Nganvongpanit K. Poommouang A, et al. Sci Rep. 2021 Jun 2;11(1):11624. doi: 10.1038/s41598-021-90947-4. Sci Rep. 2021. PMID: 34078973 Free PMC article. - Bayesian Phylogeographic Analysis Incorporating Predictors and Individual Travel Histories in BEAST.
Hong SL, Lemey P, Suchard MA, Baele G. Hong SL, et al. Curr Protoc. 2021 Apr;1(4):e98. doi: 10.1002/cpz1.98. Curr Protoc. 2021. PMID: 33836121 Free PMC article. - The Asian plethodontid salamander preserves historical genetic imprints of recent northern expansion.
Jeon JY, Jung JH, Suk HY, Lee H, Min MS. Jeon JY, et al. Sci Rep. 2021 Apr 28;11(1):9193. doi: 10.1038/s41598-021-88238-z. Sci Rep. 2021. PMID: 33911092 Free PMC article. - Genetic patterns reveal geographic drivers of divergence in silvereyes (Zosterops lateralis).
Radu A, Dudgeon C, Clegg SM, Foster Y, Levengood AL, Sendell-Price AT, Townsend KA, Potvin DA. Radu A, et al. Sci Rep. 2024 Sep 3;14(1):20426. doi: 10.1038/s41598-024-71364-9. Sci Rep. 2024. PMID: 39227633 Free PMC article. - Cooperation between passive and active silicon transporters clarifies the ecophysiology and evolution of biosilicification in sponges.
Maldonado M, López-Acosta M, Beazley L, Kenchington E, Koutsouveli V, Riesgo A. Maldonado M, et al. Sci Adv. 2020 Jul 8;6(28):eaba9322. doi: 10.1126/sciadv.aba9322. eCollection 2020 Jul. Sci Adv. 2020. PMID: 32832609 Free PMC article.
References
- Beerli P. (2006). Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22:341–345. - PubMed
- Drummond, A.J.,, Rambaut, A.,, Shapiro, B.,, Pybus, O.G. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185–1192. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases