Eosinophilic oesophagitis endotype classification by molecular, clinical, and histopathological analyses: a cross-sectional study - PubMed (original) (raw)
Observational Study
doi: 10.1016/S2468-1253(18)30096-7. Epub 2018 May 3.
Ting Wen 1, Seema S Aceves 2, J Pablo Abonia 1, Dan Atkins 3, Peter A Bonis 4, Julie M Caldwell 1, Kelley E Capocelli 5, Christina L Carpenter 6, Margaret H Collins 7, Evan S Dellon 8, Michael D Eby 1, Nirmala Gonsalves 9, Sandeep K Gupta 10, Gary W Falk 11, Ikuo Hirano 9, Paul Menard-Katcher 12, Jonathan T Kuhl 1, Jeffrey P Krischer 6, John Leung 4, Vincent A Mukkada 13, Jonathan M Spergel 14, Michael P Trimarchi 1, Guang-Yu Yang 15, Nives Zimmermann 1, Glenn T Furuta 16, Marc E Rothenberg 17; Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR)
Affiliations
- PMID: 29730081
- PMCID: PMC5997568
- DOI: 10.1016/S2468-1253(18)30096-7
Observational Study
Eosinophilic oesophagitis endotype classification by molecular, clinical, and histopathological analyses: a cross-sectional study
Tetsuo Shoda et al. Lancet Gastroenterol Hepatol. 2018 Jul.
Abstract
Background: Eosinophilic oesophagitis is understood in terms of quantifiable histological, endoscopic, and molecular features. Data are scant for inter-relations of these features and their potential to identify distinct disease endotypes. We aimed to identify clinical-pathological correlations between endoscopic and histological disease variables by transcription profiling of the oesophagus of patients with eosinophilic oesophagitis of varying severity and disease activity states.
Methods: We did a cross-sectional study across ten hospital sites in the USA associated with the Consortium of Eosinophilic Gastrointestinal Disease Researchers. We analysed oesophageal biopsy specimens taken from paediatric and adult patients with eosinophilic oesophagitis (discovery cohort), using the eosinophilic oesophagitis diagnostic panel (EDP), a set of 96 informative transcripts. Histological and endoscopic features were assessed by quantification of oesophageal eosinophils and use of the eosinophilic oesophagitis histology scoring system (HSS) and the eosinophilic oesophagitis endoscopic reference score (EREFS). Associations among the various histological, endoscopic, and molecular features were analysed by Spearman correlation. Results were replicated in a biologically independent, single-centre, validation cohort of patients with active eosinophilic oesophagitis.
Findings: The discovery cohort contained 185 samples and the validation cohort comprised 100 specimens. In the discovery cohort, EDP showed intersite consistency, significant correlation with oesophageal eosinophils (p<0·0001), and similar findings between paediatric and adult patients. Of eight HSS domains, basal zone hyperplasia correlated with the EDP (median Spearman ρ 0·47 [IQR 0·36-0·60]). Of five EREFS features, distal furrows correlated with the EDP (median Spearman ρ 0·42 [0·32-0·50]). By analysing active eosinophilic oesophagitis in the discovery cohort, the EDP identified three clusters associated with distinct endotypes (termed EoEe1-3) despite similar eosinophil levels. EoEe1 was associated with a normal-appearing oesophagus (risk ratio [RR] 3·27, 95% CI 1·04-10·27; p=0·0443), an inverse association with a history of oesophageal dilation (0·27, 0·09-0·82; p=0·0105) and showed relatively mild histological, endoscopic, and molecular changes. EoEe2 showed an inflammatory and steroid-refractory phenotype (RR 2·77, 95% CI 1·11-6·95; p=0·0376) and had the highest expression of inflammatory cytokines and steroid-responding genes. EoEe3 was associated with a narrow-calibre oesophagus (RR 7·98, 95% CI 1·84-34·64; p=0·0013) and adult onset (2·22, 1·19-4·12; p=0·0155), and showed the highest degree of endoscopic and histological severity and the lowest expression of epithelial differentiation genes. These endotypes were replicated in the validation cohort by clustering and with an eosinophilic oesophagitis endotype-prediction algorithm.
Interpretation: Our new disease classification stratifies patients with eosinophilic oesophagitis into subgroups with potential clinical and therapeutic significance and provides a framework for a precision medicine approach to eosinophilic oesophagitis.
Funding: National Institutes of Health.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflicts of Interest. M.E.R. is a consultant for Pulm One, Spoon Guru, Celgene, Shire, Astra Zeneca, Glaxosmithkline, Allakos, Adare, Regeneron and Novartis and has an equity interest in Pulm One, Spoon Guru, Celgene and Immune Pharmaceuticals and royalties from reslizumab (Teva Pharmaceuticals). M.E.R. is an inventor of patents, owned by Cincinnati Children’s. G.W.F. has received research support from Celgene/Receptos, Regeneron, Shire and Adare. M.H.C. is a consultant for Shire, Regeneron, Receptos and Adare and has received research funding from Shire, Regeneron and Receptos. S.K.G. is a consultant for Abbott, Allakos, QOL, Meritage, and Receptos and receives research support from Shire. V.A.M. is a consultant for Shire and has received research funding from Shire. N.G. is a consultant for Allakos. E.S.D. is a consultant for Adare, Alivio, Allakos, Banner, Enumeral, GSK, Receptos/Celegene, Regeneron and Shire, has received research funding from Adare, Meritage, Miraca, Nutricia, Receptos/Celgene and Shire, and has received educational grants from Banner and Holoclara. S.S.A. is a consultant for Regeneron, is an inventor of oral viscous budesonide, patented by UCSD and licensed by Shire, and has research funding from Ferring Research Institute. J.M.S. is a consultant for Regeneron and DBV Technology, and his research is supported by NIH, EATS foundation, AImmune Therapeutics, FARE and DBV Technology. I.H. is a consultant for Regeneron, Receptos, Shire, Allakos and Adare and has received research funding from Regeneron, Receptos, Shire and Adare. G.T.F. is a consultant for Shire and a co-founder of EnteroTrack. All other authors declare they have no competing interests.
Figures
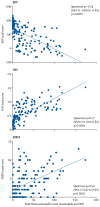
Figure 1
Associations between the peak esophageal eosinophil counts in EoE and diagnostic platforms. A linear correlation is seen between the peak esophageal eosinophils/HPF and the total score from the EDP (left), HSS (middle) and EREFS (right), with Spearman ρ, 95% confidence intervals (CI), and p-values shown. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. HSS=EoE histology scoring system. EREFS=EoE endoscopic reference score. HPF=high-power field.
Figure 2
Associations between the EDP and the HSS domains and EREFS features. Spearman correlation analysis between the gene levels on the EDP and HSS domains (left) and the EREFS features (right), using the absolute value to account for differences in the direction of the effect across genes. Statistical significance was calculated with the Kruskal-Wallis test and Dunn’s post-hoc test. *p<0·0001 vs EA, ESL, DEC, DIS, SEA and LPF. †p<0·05 vs edema, exudates, rings and stricture. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. HSS=EoE histology scoring system. EREFS=EoE endoscopic reference score. EI=eosinophilic inflammation. BZH=basal zone hyperplasia. EA=eosinophilic abscess. ESL=eosinophilic surface layering. DIS=dilated intercellular spaces. SEA=surface epithelial alteration. DEC=dyskeratotic epithelial cells. LPF=lamina propria fibers.
Figure 3
Clustering analysis of the active EoE group in the discovery cohort. (A) Consensus cumulative distribution function (CDF) with increasing number of clusters (_k_2 to _k_12). (B) Unsupervised consensus clustering of the active EoE showed optimal partitioning to 3 clusters (endotypes). (C) Comparison of esophageal transcriptomes by endotype. Heat maps were generated on the basis of the 95 EDP genes. (D) A 3-dimensional plot containing sample points from the 3 endotypes was derived from principal component analysis (PCA) of the entities demonstrated in the heat map to visualize the geometric distance between any given samples. (E) Venn diagrams comparing the number of genes identified as differentially expressed genes (adjusted p<0·05 and 2-fold change) that characterize the 3 endotypes. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. EoEe=eosinophilic esophagitis endotype.
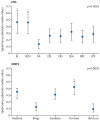
Figure 4
The clinical features of each EoE endotype. (A) Comparison of each EoE endotype by diagnostic platform. Peak esophageal eosinophil counts (upper left), EoE score from EDP (upper right), HSS scores (lower left) and EREFS score (lower right) in each EoE endotype. Data are medians (IQR). Each dot represents an individual subject. Statistical significance was calculated with the Kruskal-Wallis test and Dunn’s post-hoc test. * p<0·05, **p<0·01, ***p<0·001, ****p<0·0001, vs EoEe1. (B) Comparison of each HSS domain in each EoE endotype (upper) and each EREFS feature in each EoE endotype (lower). Data are means ± SEM. (C) Summary of significant associations for each endotype. The dashed line in the forest plots indicates a risk ratio of 1. (D) Multiple correspondence analyses plot of the relationships between clinical phenotypes and endotypes. Distance between variables (phenotype and endotype) indicates the approximate relationship between variables. The distance between variables is inversely proportional to the strength of the relationship. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. EoEe=eosinophilic esophagitis endotype. HSS=EoE histology scoring system, EREFS=EoE endoscopic reference score. HPF=high-power field. RR=risk ratio. CI= confidence interval.
Figure 5
EoE endotype prediction based on machine learning with high accuracy. (A) Stepwise discriminant analysis shows the 15 strongest discriminatory genes for cluster assignment. (B) Canonical plot in which subjects are plotted in a 2-dimensional space. Each dot represents an individual subject. A 95% confidence level ellipse (inner) and an ellipse denoting a 50% contour (outer) are plotted for each group. The flow of the analysis was graphed in the lower table. The diagnostic accuracy was summarized in the right table. EDP=EoE diagnostic panel. EoE=eosinophilic esophagitis. EoEe=eosinophilic esophagitis endotype. PPV=positive predictive value. NPV=negative predictive value.
Comment in
- Endophenotyping eosinophilic oesophagitis: a new era for management?
Keely S, Talley NJ. Keely S, et al. Lancet Gastroenterol Hepatol. 2018 Jul;3(7):449-450. doi: 10.1016/S2468-1253(18)30134-1. Epub 2018 May 3. Lancet Gastroenterol Hepatol. 2018. PMID: 29730082 No abstract available.
Similar articles
- One-food versus six-food elimination diet therapy for the treatment of eosinophilic oesophagitis: a multicentre, randomised, open-label trial.
Kliewer KL, Gonsalves N, Dellon ES, Katzka DA, Abonia JP, Aceves SS, Arva NC, Besse JA, Bonis PA, Caldwell JM, Capocelli KE, Chehade M, Cianferoni A, Collins MH, Falk GW, Gupta SK, Hirano I, Krischer JP, Leung J, Martin LJ, Menard-Katcher P, Mukkada VA, Peterson KA, Shoda T, Rudman Spergel AK, Spergel JM, Yang GY, Zhang X, Furuta GT, Rothenberg ME. Kliewer KL, et al. Lancet Gastroenterol Hepatol. 2023 May;8(5):408-421. doi: 10.1016/S2468-1253(23)00012-2. Epub 2023 Feb 28. Lancet Gastroenterol Hepatol. 2023. PMID: 36863390 Free PMC article. Clinical Trial. - Endoscopic appearance and location dictate diagnostic yield of biopsies in eosinophilic oesophagitis.
Salek J, Clayton F, Vinson L, Saffari H, Pease LF 3rd, Boynton K, Fang J, Cox K, Peterson KA. Salek J, et al. Aliment Pharmacol Ther. 2015 Jun;41(12):1288-95. doi: 10.1111/apt.13201. Epub 2015 Apr 21. Aliment Pharmacol Ther. 2015. PMID: 25898774 - The Endoscopic Reference Score shows modest accuracy to predict either clinical or histological activity in adult patients with eosinophilic oesophagitis.
Rodríguez-Sánchez J, Barrio-Andrés J, Nantes Castillejo O, Valdivieso-Cortazar E, Pérez-Martínez I, Boumidi A, Olmos-Jérez JA, Payeras-Llodra G, Alcaide-Suarez N, Ruiz-Rebollo L, Madrigal-Rubiales B, Gonzalez-Obeso E, de la Santa Belda E, López Viedma B, Molina-Infante J. Rodríguez-Sánchez J, et al. Aliment Pharmacol Ther. 2017 Jan;45(2):300-309. doi: 10.1111/apt.13845. Epub 2016 Nov 20. Aliment Pharmacol Ther. 2017. PMID: 27868216 - Review article: oesophageal dilation in adults with eosinophilic oesophagitis.
Bohm ME, Richter JE. Bohm ME, et al. Aliment Pharmacol Ther. 2011 Apr;33(7):748-57. doi: 10.1111/j.1365-2036.2011.04593.x. Epub 2011 Feb 14. Aliment Pharmacol Ther. 2011. PMID: 21320137 Review. - Overview of eosinophilic oesophagitis.
Attwood SE. Attwood SE. Br J Hosp Med (Lond). 2019 Mar 2;80(3):132-138. doi: 10.12968/hmed.2019.80.3.132. Br J Hosp Med (Lond). 2019. PMID: 30860925 Review.
Cited by
- Real world treatment patterns in patients with eosinophilic esophagitis in Japan.
Sawada A, Ihara Y, Imai T, Tanaka F, Fujiwara Y. Sawada A, et al. Sci Rep. 2024 Nov 11;14(1):27490. doi: 10.1038/s41598-024-78868-4. Sci Rep. 2024. PMID: 39528636 Free PMC article. - Advances in omics data for eosinophilic esophagitis: moving towards multi-omics analyses.
Matsuyama K, Yamada S, Sato H, Zhan J, Shoda T. Matsuyama K, et al. J Gastroenterol. 2024 Nov;59(11):963-978. doi: 10.1007/s00535-024-02151-6. Epub 2024 Sep 19. J Gastroenterol. 2024. PMID: 39297956 Free PMC article. Review. - Examining the Role of Type 2 Inflammation in Eosinophilic Esophagitis.
Chehade M, Falk GW, Aceves S, Lee JK, Mehta V, Leung J, Shumel B, Jacob-Nara JA, Deniz Y, Rowe PJ, Cunoosamy D, Khodzhayev A. Chehade M, et al. Gastro Hep Adv. 2022 May 21;1(5):720-732. doi: 10.1016/j.gastha.2022.05.004. eCollection 2022. Gastro Hep Adv. 2022. PMID: 39131849 Free PMC article. Review. - Advances and ongoing challenges in eosinophilic gastrointestinal disorders presented at the CEGIR/TIGERs Symposium at the 2024 American Academy of Allergy, Asthma & Immunology meeting.
Wright BL, Abonia JP, Abud EM, Aceves SS, Ackerman SJ, Braskett M, Chang JW, Chehade M, Constantine GM, Davis CM, Dellon ES, Doyle AD, Durban R, Hill DA, Jensen ET, Kewalramani A, Khoury P, Klion AD, Kottyan L, Kuang FL, McGowan EC, Ruffner MA, Spencer LA, Spergel JM, Uchida AM, Wechsler JB, Pesek RD. Wright BL, et al. J Allergy Clin Immunol. 2024 Oct;154(4):882-892. doi: 10.1016/j.jaci.2024.07.022. Epub 2024 Aug 5. J Allergy Clin Immunol. 2024. PMID: 39111348 Review. - Losartan Treatment Reduces Esophageal Eosinophilic Inflammation in a Subset of Eosinophilic Esophagitis.
Abonia JP, Rudman Spergel AK, Hirano I, Shoda T, Zhang X, Martin LJ, Mukkada VA, Putnam PE, Blacklidge M, Neilson D, Collins MH, Yang GY, Capocelli KE, Foote H, Eby M, Dong S, Aceves SS, Rothenberg ME; Consortium of Eosinophilic Gastrointestinal Disease Researchers. Abonia JP, et al. J Allergy Clin Immunol Pract. 2024 Sep;12(9):2427-2438.e3. doi: 10.1016/j.jaip.2024.07.011. Epub 2024 Jul 25. J Allergy Clin Immunol Pract. 2024. PMID: 39059581 Clinical Trial.
References
- Simon D, Cianferoni A, Spergel JM, et al. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy. 2016;71(5):611–20. - PubMed
- Dellon ES, Gonsalves N, Hirano I, et al. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE) Am J Gastroenterol. 2013;108(5):679–92. - PubMed
- Warners MJ, Hindryckx P, Levesque BG, et al. Systematic Review: Disease Activity Indices in Eosinophilic Esophagitis. Am J Gastroenterol. 2017;112(11):1658–69. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical