Phosphorylation-Mediated Clearance of Amyloid-like Assemblies in Meiosis - PubMed (original) (raw)
Phosphorylation-Mediated Clearance of Amyloid-like Assemblies in Meiosis
Kayla Carpenter et al. Dev Cell. 2018.
Abstract
Amyloids are fibrous protein assemblies that are often described as irreversible and intrinsically pathogenic. However, yeast cells employ amyloid-like assemblies of the RNA-binding protein Rim4 to control translation during meiosis. Here, we show that multi-site phosphorylation of Rim4 is critical for its regulated disassembly and degradation and that failure to clear Rim4 assemblies interferes with meiotic progression. Furthermore, we identify the protein kinase Ime2 to bring about Rim4 clearance via phosphorylation of Rim4's intrinsically disordered region. Rim4 phosphorylation leads to reversal of its amyloid-like properties and degradation by the proteasome. Our data support a model in which a threshold amount of phosphorylation, rather than modification of critical residues, is required for Rim4 clearance. Our results further demonstrate that at least some amyloid-like assemblies are not as irreversible as previously thought. We propose that the natural pathways by which cells process these structures could be deployed to act on disease-related amyloids.
Keywords: RNA-binding proteins; amyloid-like assemblies; gametogenesis; meiosis; neurodegeneration; phosphorylation; translation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
All authors declare no competing interests
Figures
Figure 1. Rim4 is hyperphosphorylated during meiosis
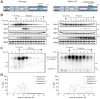
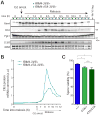
(A) Rim4 immunoprecipitation (IP) from meiotic cells. RIM4-3V5, pGAL-NDT80, GAL4.ER (B48) cells were induced to sporulate at 30°C. At 6 hours when cells had arrested in G2 due to the lack of Ndt80, cells were released from the G2 block by the addition of 1 μM β-estradiol. Lysates were prepared from G2-arrested cells (6 hour) and at 7, 8, 9 and 10 hours (post-release). G2-arrested pGAL-NDT80, GAL4.ER (A15055) was included as a no-tag control. IP was conducted under denaturing conditions using anti-V5 agarose beads. Bound factors were released by boiling in SDS-loading buffer and resolved by SDS-PAGE. Imperial blue-stained gels of the IPs are shown. Excised Rim4 bands were analyzed by mass spectrometry. (B) Diagram of Rim4 protein and time course analysis of phosphorylation sites in meiosis by mass spectrometry. The left diagram shows the domain structure of Rim4 with all serine and threonine residues marked in red and blue, IDRs shaded in purple, and the prion-like domain shaded in green. Phosphorylated residues are denoted in black. The right diagrams show time course mapping of each phosphorylation site (phosphoserine in red and phosphothreonine in blue). Regions of no mass spectrometry coverage are shown as yellow bars adjacent to the diagrams. (C, D) Phosphorylation of the Rim4 C-terminal IDR is required for its clearance. RIM4-3V5 (wild type, B48) and RIM4-47A-3V5 (A38075) strains harboring pGAL-NDT80, GAL4.ER, and CLB3-3HA were induced to sporulate 30°C. After 6 hours, cells were released from the G2 arrest. (C) Single-cell Rim4 levels in metaphase I, anaphase I, metaphase II, and anaphase II were determined by immunofluorescence (IF) (n = 50 cells for each meiotic stage). (D) The images show Rim4 protein (red), tubulin (green) and DNA (DAPI) in representative anaphase I and anaphase II cells.
Figure 2. Phosphorylation-driven clearance of Rim4 assemblies is required for timely meiotic progression
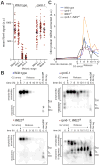
Left panel: wild type, right panel: RIM4-47A (A) Diagrams of Rim4. The region mutated in the RIM4-47A allele is bracketed. This region harbors 47 serine and threonine residues mutated to alanine. (B – D) RIM4-47A cells exhibit defective CLB3 translation, failure to clear SDS-resistant assemblies, and delayed meiotic progression. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and either RIM4-3V5 (wild type, B48) or RIM4-47A-3V5 (A38075) were induced to sporulate 30°C. After 6 ho urs, cells were released from the G2 arrest. (B) Rim4-3V5, Clb3-3HA, and Pgk1 (loading control) protein and CLB3 mRNA and rRNA (loading control) levels were determined at the indicated times following β-estradiol addition. (C) Rim4’s ability to form SDS-resistant amyloid-like assemblies was analyzed by SDD-AGE as in (Halfmann and Lindquist, 2008) with minor modifications (see experimental procedures). (D) The percentage (n = 100 cells for each time point) of metaphase I, anaphase I, metaphase II, and anaphase II cells was determined by tubulin IF and DAPI staining.
Figure 3. RIM4-47A is a dominant gain-of-function allele
(A, B) A single copy of RIM4-47A is sufficient to cause CLB3 translational control defects. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and either RIM4-3V5/+ (light blue, B119) or RIM4-47A-3V5/+ (light green, B120) were induced to sporulate at 30°C. After 6 hours, cells were released from the G2 arrest. (A) Rim4-3V5, Clb3-3HA, and Pgk1 (loading control) protein and CLB3 mRNA and rRNA (loading control) levels were determined at the indicated times following release from G2 arrest. (B) Clb3 protein levels corrected for CLB3 mRNA levels are plotted on the y-axis and time in meiosis is plotted on the x-axis. Analysis of Rim4 SDS-resistant assemblies and meiotic progression is shown in Figure S3. (C) RIM4-47A cells exhibit reduced spore viability. Strains harboring RIM4-3V5/RIM4-3V5 (blue, B123) RIM4-47A-3V5/RIM4-3V5 (light green, B149), and RIM4-47A/RIM4-47A (green, B126) were sporulated on plates. Tetrads (n = 7 plates of 20 tetrads for each genotype) were dissected on YPD plates and spores were grown for 2 days. Shown is the percent viable spores. A Mann-Whitney test was used to statistically compare the means. * = P < 0.05
Figure 4. Rim4 clearance is regulated by multi-site phosphorylation
(A) Diagram of Rim4 with serine and threonine residues denoted in red and blue, respectively. The IDRs are shaded in purple. Mutations contained in each RIM4 phospho-mutant allele are shaded in red or orange (S/T → A and S/T → E respectively). Translational control phenotypes (determined from (B)) are indicated on the right. (B) CLB3 translational control analysis of RIM4 phospho-mutant alleles. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and one of the following: RIM4-3V5 (wild type, blue, B48), rim4-99A-3V5 (red, A30860), RIM4-52A-3V5 (green, B67), RIM4-47A-3V5 (green, A38075), RIM4-36A-3V5 (green, B70), RIM4-27A-3V5 (light green, B152), RIM4-25A-3V5 (light green, B155), RIM4-21A-3V5 (light green, B129), rim4-16A-3V5 (blue, B73), or rim4-10A-3V5 (blue, A38072) were induced to sporulate 30°C. After 6 hours, cells were released from the G 2 arrest. Four strains were run per experiment, each containing a RIM4-3V5 (wild-type) control and three mutant strains. Clb3 protein levels corrected for CLB3 mRNA level are plotted on the y-axis and time in meiosis is plotted on the x-axis. Western and Northern blot source data is shown in Figure S4. (C–E) rim4-99A and rim4-45E are loss of function alleles. (C, D) Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and one of the following: RIM4-3V5 (wild type, B48), rim4Δ (B343), RIM4-47A-3V5 (A38075), rim4-99A-3V5 (A30860), and rim4-45E-3V5 (B346) were induced to sporulate at 30°C. After 6 hours, cells were released from the G2 arrest. Meiotic progression was analyzed by tubulin IF (n = 100 cells for each time point). (E) Rim4 SDS-resistant assemblies were analyzed by SDD-AGE in RIM4-3V5 (wild type) and rim4-45E-3V5 cells.
Figure 5. Ime2 regulates Rim4 clearance via phosphorylation of the Rim4 C-terminal IDR
(A, B) Epistatic analysis of IME2st and RIM4-47A translational control phenotypes. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and either RIM4-3V5 (wild type, blue, B48), IME2st (yellow, A33024), RIM4-47A-3V5 (green, A38075), or IME2st; RIM4-47A-3V5 (black, B5) were induced to sporulate at 30°C. After 6 hours, cells were releas ed from the G2 arrest. (A) Rim4-3V5, Clb3-3HA, and Pgk1 (loading control) protein and CLB3 mRNA and rRNA (loading control) levels were determined at the indicated times. (B) Clb3 protein levels corrected for CLB3 mRNA levels are plotted on the y-axis and time in meiosis is plotted on the x-axis. Western and Northern blot data for later time points is shown in Figure S5. (C) Rim4 SDS-resistant assemblies were analyzed by SDD-AGE. (D) Meiotic progression was analyzed by tubulin IF (n = 100 cells for each time point).
Figure 6. Rim4 assemblies are disassembled prior to proteasomal degradation
(A) The proteasome is critical for Rim4 clearance. Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, and RIM4-3V5 and either RPN6 (wild type) or rpn6-1 were induced to sporulate at 30°C. After 6 hours, cells were released from the G2 arrest. Single-cell Rim4 levels in metaphase I, anaphase I, metaphase II, and anaphase II were determined by immunofluorescence (IF) (n = 48 for each meiotic stage except rpn6-1 metaphase II in which n = 26 and anaphase II in which n = 0). CLB3 translational control and meiotic progression analysis is shown in Figures S6A, S6B. (B, C) Strains harboring pGAL-NDT80, GAL4.ER, CLB3-3HA, RIM4-3V5 and either RPN6 (wild type, blue, B48), rpn6-1 (purple, B207), IME2st (yellow, A33024), or rpn6-1; IME2st (black, B251) were grown as in (A). (B) Rim4 SDS-resistant assemblies were analyzed by SDD-AGE. (C) CLB3 translational control analysis. Clb3 protein levels corrected for CLB3 mRNA levels are plotted on the y-axis and time in meiosis is plotted on the x-axis. Quantification of mRNA-corrected Clb3 protein levels is shown. Source data and meiotic progression analysis is shown in Figures S6C, S6D.
Figure 7. Model of Rim4 clearance
Comment in
- Phosphorylation Leads the Way for Protein Aggregate Disassembly.
Boeynaems S, Gitler AD. Boeynaems S, et al. Dev Cell. 2018 May 7;45(3):279-281. doi: 10.1016/j.devcel.2018.04.017. Dev Cell. 2018. PMID: 29738705
Similar articles
- Clearance of an amyloid-like translational repressor is governed by 14-3-3 proteins.
Herod SG, Dyatel A, Hodapp S, Jovanovic M, Berchowitz LE. Herod SG, et al. Cell Rep. 2022 May 3;39(5):110753. doi: 10.1016/j.celrep.2022.110753. Cell Rep. 2022. PMID: 35508136 Free PMC article. - Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis.
Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Berchowitz LE, et al. Cell. 2015 Oct 8;163(2):406-18. doi: 10.1016/j.cell.2015.08.060. Epub 2015 Sep 24. Cell. 2015. PMID: 26411291 Free PMC article. - Assembly and function of the amyloid-like translational repressor Rim4 is coupled with nutrient conditions.
Ottoz DS, Tang LC, Dyatel AE, Jovanovic M, Berchowitz LE. Ottoz DS, et al. EMBO J. 2023 Dec 1;42(23):e113332. doi: 10.15252/embj.2022113332. Epub 2023 Nov 3. EMBO J. 2023. PMID: 37921330 Free PMC article. - The Ime2 protein kinase family in fungi: more duties than just meiosis.
Irniger S. Irniger S. Mol Microbiol. 2011 Apr;80(1):1-13. doi: 10.1111/j.1365-2958.2011.07575.x. Epub 2011 Mar 1. Mol Microbiol. 2011. PMID: 21306447 Review. - A reversible liquid drop aggregation controls glucose response in yeast.
Simpson-Lavy K, Kupiec M. Simpson-Lavy K, et al. Curr Genet. 2018 Aug;64(4):785-788. doi: 10.1007/s00294-018-0805-0. Epub 2018 Jan 10. Curr Genet. 2018. PMID: 29322248 Review.
Cited by
- Phospho-Regulation of Meiotic Prophase.
Kar FM, Hochwagen A. Kar FM, et al. Front Cell Dev Biol. 2021 Apr 13;9:667073. doi: 10.3389/fcell.2021.667073. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33928091 Free PMC article. Review. - Kar4 is required for the normal pattern of meiotic gene expression.
Park ZM, Remillard M, Belnap E, Rose MD. Park ZM, et al. PLoS Genet. 2023 Aug 28;19(8):e1010898. doi: 10.1371/journal.pgen.1010898. eCollection 2023 Aug. PLoS Genet. 2023. PMID: 37639444 Free PMC article. - Spo13/MEIKIN ensures a Two-Division meiosis by preventing the activation of APC/CAma1 at meiosis I.
Rojas J, Oz T, Jonak K, Lyzak O, Massaad V, Biriuk O, Zachariae W. Rojas J, et al. EMBO J. 2023 Oct 16;42(20):e114288. doi: 10.15252/embj.2023114288. Epub 2023 Sep 20. EMBO J. 2023. PMID: 37728253 Free PMC article. - Gametogenesis: Exploring an Endogenous Rejuvenation Program to Understand Cellular Aging and Quality Control.
Sing TL, Brar GA, Ünal E. Sing TL, et al. Annu Rev Genet. 2022 Nov 30;56:89-112. doi: 10.1146/annurev-genet-080320-025104. Epub 2022 Jul 25. Annu Rev Genet. 2022. PMID: 35878627 Free PMC article. Review. - RNA binding protein BOULE forms aggregates in mammalian testis.
Su Y, Guo X, Zang M, Xie Z, Zhao T, Xu EY. Su Y, et al. J Biomed Res. 2022 Jun 28;36(4):255-268. doi: 10.7555/JBR.36.20220072. J Biomed Res. 2022. PMID: 35965435 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases