MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches - PubMed (original) (raw)
Review
MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches
Nicola Amodio et al. J Hematol Oncol. 2018.
Abstract
The deeper understanding of non-coding RNAs has recently changed the dogma of molecular biology assuming protein-coding genes as unique functional biological effectors, while non-coding genes as junk material of doubtful significance. In the last decade, an exciting boom of experimental research has brought to light the pivotal biological functions of long non-coding RNAs (lncRNAs), representing more than the half of the whole non-coding transcriptome, along with their dysregulation in many diseases, including cancer.In this review, we summarize the emerging insights on lncRNA expression and functional role in cancer, focusing on the evolutionary conserved and abundantly expressed metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) that currently represents the best characterized lncRNA. Altogether, literature data indicate aberrant expression and dysregulated activity of MALAT1 in human malignancies and envision MALAT1 targeting as a novel treatment strategy against cancer.
Keywords: Epigenetics; Experimental therapeutics; Long non-coding RNA; MALAT1; Non-coding RNA; lncRNA.
Conflict of interest statement
Ethics approval and consent to participate
This is not applicable for this review.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
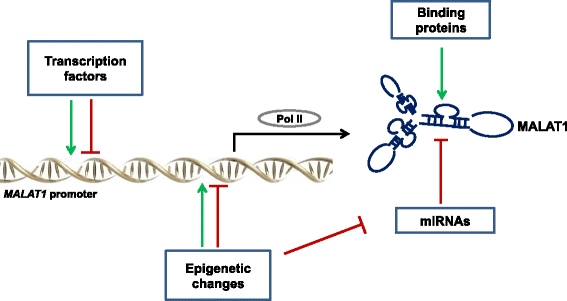
Fig. 1
Mechanisms of MALAT1 regulation. MALAT1 expression can be positively or negatively affected by transcription factors, epigenetic changes (histone or CpG methylation/demethylation), miRNAs and binding proteins stabilizing the triple helix, such as METTL16
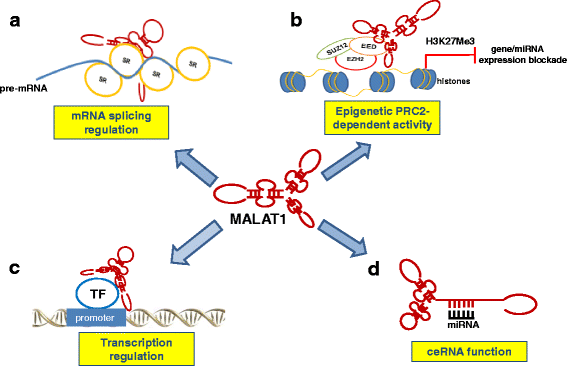
Fig. 2
MALAT1 functions in human cancer. a: MALAT1 can affect mRNA transcription by regulating splicing of pre-mRNAs through interacting with and regulating phosphorylation of serine- and arginine-rich (SR) proteins into nuclear speckles. b: MALAT1 interacts with PRC2 components EZH2, SUZ12, and EED and reduces target gene or miRNA expression by promoting trimethylation of histone H3 at lysine 27 (H3K27me3). c: MALAT1 can affect mRNA transcription by facilitating transcription factor (TF) binding to promoter of target genes. d: MALAT1 can sequestrate miRNAs acting as a sponge, thus activating the expression of miRNA targets
Similar articles
- The long noncoding RNA Malat1: Its physiological and pathophysiological functions.
Zhang X, Hamblin MH, Yin KJ. Zhang X, et al. RNA Biol. 2017 Dec 2;14(12):1705-1714. doi: 10.1080/15476286.2017.1358347. Epub 2017 Oct 6. RNA Biol. 2017. PMID: 28837398 Free PMC article. Review. - Targeting long non-coding RNAs in cancers: progress and prospects.
Li CH, Chen Y. Li CH, et al. Int J Biochem Cell Biol. 2013 Aug;45(8):1895-910. doi: 10.1016/j.biocel.2013.05.030. Epub 2013 Jun 4. Int J Biochem Cell Biol. 2013. PMID: 23748105 Review. - Metastasis Associated Lung Adenocarcinoma Transcript 1: An update on expression pattern and functions in carcinogenesis.
Ghafouri-Fard S, Ashrafi Hafez A, Taheri M. Ghafouri-Fard S, et al. Exp Mol Pathol. 2020 Feb;112:104330. doi: 10.1016/j.yexmp.2019.104330. Epub 2019 Nov 9. Exp Mol Pathol. 2020. PMID: 31712117 - LncRNA MALAT1-related signaling pathways in osteosarcoma.
Farzaneh M, Najafi S, Anbiyaee O, Azizidoost S, Khoshnam SE. Farzaneh M, et al. Clin Transl Oncol. 2023 Jan;25(1):21-32. doi: 10.1007/s12094-022-02876-x. Epub 2022 Jul 6. Clin Transl Oncol. 2023. PMID: 35790599 Review. - Long non-coding RNA MALAT1 functions as miR-1 sponge to regulate Connexin 43-mediated ossification of the posterior longitudinal ligament.
Yuan X, Guo Y, Chen D, Luo Y, Chen D, Miao J, Chen Y. Yuan X, et al. Bone. 2019 Oct;127:305-314. doi: 10.1016/j.bone.2019.06.019. Epub 2019 Jul 4. Bone. 2019. PMID: 31280017
Cited by
- Long Non-Coding RNAs as Strategic Molecules to Augment the Radiation Therapy in Esophageal Squamous Cell Carcinoma.
Sharma U, Barwal TS, Acharya V, Singh K, Rana MK, Singh SK, Prakash H, Bishayee A, Jain A. Sharma U, et al. Int J Mol Sci. 2020 Sep 16;21(18):6787. doi: 10.3390/ijms21186787. Int J Mol Sci. 2020. PMID: 32947897 Free PMC article. - Non-Coding RNA-Dependent Regulation of Mitochondrial Dynamics in Cancer Pathophysiology.
Gallo Cantafio ME, Torcasio R, Viglietto G, Amodio N. Gallo Cantafio ME, et al. Noncoding RNA. 2023 Feb 20;9(1):16. doi: 10.3390/ncrna9010016. Noncoding RNA. 2023. PMID: 36827549 Free PMC article. Review. - IL-8 Secreted from M2 Macrophages Promoted Prostate Tumorigenesis via STAT3/MALAT1 Pathway.
Zheng T, Ma G, Tang M, Li Z, Xu R. Zheng T, et al. Int J Mol Sci. 2018 Dec 27;20(1):98. doi: 10.3390/ijms20010098. Int J Mol Sci. 2018. PMID: 30591689 Free PMC article. - Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter.
Qu D, Sun WW, Li L, Ma L, Sun L, Jin X, Li T, Hou W, Wang JH. Qu D, et al. Nucleic Acids Res. 2019 Apr 8;47(6):3013-3027. doi: 10.1093/nar/gkz117. Nucleic Acids Res. 2019. PMID: 30788509 Free PMC article. - Probing the function of long noncoding RNAs in the nucleus.
Akkipeddi SMK, Velleca AJ, Carone DM. Akkipeddi SMK, et al. Chromosome Res. 2020 Mar;28(1):87-110. doi: 10.1007/s10577-019-09625-x. Epub 2020 Feb 6. Chromosome Res. 2020. PMID: 32026224 Free PMC article. Review.
References
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. - PubMed
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. - PubMed
- Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources