Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition - PubMed (original) (raw)
Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition
Marios G Koliopoulos et al. Nat Commun. 2018.
Abstract
RIG-I is a viral RNA sensor that induces the production of type I interferon (IFN) in response to infection with a variety of viruses. Modification of RIG-I with K63-linked poly-ubiquitin chains, synthesised by TRIM25, is crucial for activation of the RIG-I/MAVS signalling pathway. TRIM25 activity is targeted by influenza A virus non-structural protein 1 (NS1) to suppress IFN production and prevent an efficient host immune response. Here we present structures of the human TRIM25 coiled-coil-PRYSPRY module and of complexes between the TRIM25 coiled-coil domain and NS1. These structures show that binding of NS1 interferes with the correct positioning of the PRYSPRY domain of TRIM25 required for substrate ubiquitination and provide a mechanistic explanation for how NS1 suppresses RIG-I ubiquitination and hence downstream signalling. In contrast, the formation of unanchored K63-linked poly-ubiquitin chains is unchanged by NS1 binding, indicating that RING dimerisation of TRIM25 is not affected by NS1.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
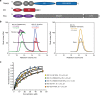
Oligomeric state of NS1 and identification of TRIM25-interacting domains. a Schematic representation of TRIM25, NS1 and RIG-I domain structures. The crystallised TRIM25 CC-PRYSPRY domain is highlighted in blue and the crystallised RBD and NS1 domains of NS1 in red. The tandem CARD construct of RIG-I used in substrate ubiquitination assays is highlighted in purple. b The effect of different mutations on the oligomeric state of NS1-FL assessed by SEC–MALLS. The traces are colour coded according to the NS1 mutant and concentration used. c Quantification of the TRIM25-CC interaction with NS1 by biolayer interferometry (BLI). The binding curves are colour-coded and _K_ds determined are listed. The error is the standard deviation (s.d.) of mean from at least three independent experiments
Fig. 2
Crystal structure of NS1-ED in complex with TRIM25-CC. a The asymmetric unit of the TRIM25-CC/NS1-ED complex containing three copies of each TRIM25-CC (blue and grey) and NS1-ED (in red, wheat and pink) monomers. b Symmetry analysis shows that NS1-ED binds to a dimeric TRIM25-CC via two distinct interfaces. The structure is shown in ribbon representation. The position of a twofold symmetry axis is shown in red. c Interface A is formed by the NS1-ED α-helix containing amino acids E95 and E96, which have previously been suggested to be important for complex formation. The insert shows a close-up view of the interface with TRIM25-CC in light and dark blue and NS1-ED in red. Interactions described in the text are highlighted by dashed lines. d Details of interface B, with TRIM25-CC in light and dark blue and NS1-ED in wheat
Fig. 3
Crystal structure of NS1-FL in complex with TRIM25-CC. a The asymmetric unit contains four molecules of each, NS1-FL (R38A/K41A/W187A) and TRIM25-CC, with electron density visible for 2 TRIM25-CC dimers (in dark and light blue), 2 NS1-FL (in red and pink) and 2 NS1-ED domains (in wheat). b NS1-FL resembles the shape of letter Y, with the RBD forming a homodimeric 6-helical structure and the effector domains extending outwards without making contact with each other or the RBDs. c Structural alignment of dimeric NS1-FL from a using the RBD for the overlap shows that the two protomers are not in identical positions, with one ED being shifted by 17 Å in relation to the other. d Structural alignment of NS1 shown in b (in red) with the other two available apo-NS1-FL structures (A/Vietnam/1203/2004(H5N1), R38A/K41A) shown in yellow (PDB: 3F5T) and (A/MN/993/1980(H6N6) R38A/K41A) shown in blue (PDB: 4OPH) reveals structural flexibility owing to the linker region (LR) of NS1
Fig. 4
Crystal structure of TRIM25 CC-PRYSPRY. a The TRIM25 CC dimer (in dark and light blue) with two PRYSPRY domains bound at either end in green cyan. There is no electron density for the linker connecting the CC and PRYSPRY domains. The distance between the N-termini of the PRYSPRY domains and C-terminus of one CC chain is indicated. b Close-up view of the interface between the CC and PRYSPRY domain. Residues described in the text are shown as ball and stick. c Ubiquitination assays of the tandem CARDs of RIG-I with TRIM25 to test the importance of Y463 and Y476 in stabilising the observed CC-PRYSPRY arrangement and promoting substrate ubiquitination. FLAG-RIG-I-2CARD and TRIM25 were co-transfected into HEK293T cells and WCLs were subjected to IP with anti-FLAG beads and immunoblotted with α-ubiquitin and α-FLAG antibodies. The asterisk indicates an unspecific band. d Comparison of the positions of the PRYSPRY domain (green cyan) and the NS1 ED (red), highlighting the changes in the position of the linker connecting the CC and PRYSPRY domain, induced by binding to NS1
Fig. 5
Inhibition of TRIM25-mediated RIG-I ubiquitination and signalling by NS1. a Effect of NS1 and mutants on the ubiquitination of 3FLAG-RIG-I-2CARD by TRIM25 in HEK293T cells. 3FLAG-RIG-I-2CARD, TRIM25 and NS1 were co-transfected into HEK293T cells and WCLs were subjected to IP with anti-FLAG beads and immunoblotted with α-ubiquitin and α-FLAG antibodies. The asterisk indicates an unspecific band. b In vitro ubiquitination assays with UBE2N/UBE2V1 and TRIM25-FL in the absence or presence of increasing amounts of NS1 supplemented with fluorescent AttoUb. Assays were carried out with NS1-ED or NS1-FL (R38A/K41A/W187A or W187A) and the reaction was monitored over 60 min. Gels were scanned with a Typhoon FLA 9500 scanner and the fluorescence converted to black and white. c 3FLAG-RIG-I-2CARD, TRIM25, and increasing amounts of NS1 WT and mutants (25, 50 and 100 ng) were co-transfected into HEK293T cells. Interferon induction by 3FLAG-RIG-I-2CARD was measured in a Luciferase-based reporter assay, where Luciferase activity is controlled by the IFN-β promoter. The mCherry plasmid was used as a control. The error bars shown are the standard deviation (s.d.) of mean from two independent experiments, each with triplicates
Fig. 6
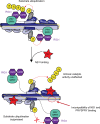
Model for suppression of substrate ubiquitination by NS1. Model of TRIM25 activity highlighting how the RING domains either side of the anti-parallel CC bind back to dimerise in an intramolecular fashion and come into close proximity of the substrate-binding PRYSPRY domain. The monomers of dimeric TRIM25 are shown in light and dark blue, with ubiquitin-loaded E2 (E2-Ub) in grey and yellow bound to each RING domain while the tandem CARDs of RIG-I (in purple) are bound to the PRYSPRY domain (in green cyan) of TRIM25. The linker connecting the CC and PRYSPRY domain is highlighted in yellow. Binding of NS1 (in red) to the CC of TRIM25 interferes with the linker position and hence correct arrangement of the PRYSPRY domain with respect to the RING domains to suppress substrate ubiquitination
Similar articles
- Human Respiratory Syncytial Virus NS 1 Targets TRIM25 to Suppress RIG-I Ubiquitination and Subsequent RIG-I-Mediated Antiviral Signaling.
Ban J, Lee NR, Lee NJ, Lee JK, Quan FS, Inn KS. Ban J, et al. Viruses. 2018 Dec 14;10(12):716. doi: 10.3390/v10120716. Viruses. 2018. PMID: 30558248 Free PMC article. - Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein.
Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. Rajsbaum R, et al. PLoS Pathog. 2012;8(11):e1003059. doi: 10.1371/journal.ppat.1003059. Epub 2012 Nov 29. PLoS Pathog. 2012. PMID: 23209422 Free PMC article. - The Human Papillomavirus E6 Oncoprotein Targets USP15 and TRIM25 To Suppress RIG-I-Mediated Innate Immune Signaling.
Chiang C, Pauli EK, Biryukov J, Feister KF, Meng M, White EA, Münger K, Howley PM, Meyers C, Gack MU. Chiang C, et al. J Virol. 2018 Feb 26;92(6):e01737-17. doi: 10.1128/JVI.01737-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263274 Free PMC article. - Ubiquitin-mediated modulation of the cytoplasmic viral RNA sensor RIG-I.
Oshiumi H, Matsumoto M, Seya T. Oshiumi H, et al. J Biochem. 2012 Jan;151(1):5-11. doi: 10.1093/jb/mvr111. Epub 2011 Sep 2. J Biochem. 2012. PMID: 21890623 Review. - TRIM proteins: another class of viral victims.
Munir M. Munir M. Sci Signal. 2010 Apr 20;3(118):jc2. doi: 10.1126/scisignal.3118jc2. Sci Signal. 2010. PMID: 20407122 Review.
Cited by
- The Small t Antigen of JC Virus Antagonizes RIG-I-Mediated Innate Immunity by Inhibiting TRIM25's RNA Binding Ability.
Chiang C, Dvorkin S, Chiang JJ, Potter RB, Gack MU. Chiang C, et al. mBio. 2021 Apr 13;12(2):e00620-21. doi: 10.1128/mBio.00620-21. mBio. 2021. PMID: 33849980 Free PMC article. - CSFV restricts necroptosis to sustain infection by inducing autophagy/mitophagy-targeted degradation of RIPK3.
Wu K, Li B, Zhang X, Fang Y, Zeng S, Hu W, Liu X, Liu X, Lu Z, Li X, Chen W, Qin Y, Zhou B, Zou L, Zhao F, Yi L, Zhao M, Fan S, Chen J. Wu K, et al. Microbiol Spectr. 2024 Jan 11;12(1):e0275823. doi: 10.1128/spectrum.02758-23. Epub 2023 Dec 15. Microbiol Spectr. 2024. PMID: 38100396 Free PMC article. - The roles and targeting options of TRIM family proteins in tumor.
Zhang Y, Zhang W, Zheng L, Guo Q. Zhang Y, et al. Front Pharmacol. 2022 Sep 30;13:999380. doi: 10.3389/fphar.2022.999380. eCollection 2022. Front Pharmacol. 2022. PMID: 36249749 Free PMC article. Review. - The influenza NS1 protein modulates RIG-I activation via a strain-specific direct interaction with the second CARD of RIG-I.
Jureka AS, Kleinpeter AB, Tipper JL, Harrod KS, Petit CM. Jureka AS, et al. J Biol Chem. 2020 Jan 24;295(4):1153-1164. doi: 10.1074/jbc.RA119.011410. Epub 2019 Dec 16. J Biol Chem. 2020. PMID: 31843969 Free PMC article. - Antiviral Gene Expression in Young and Aged Murine Lung during H1N1 and H3N2.
Harris R, Yang J, Pagan K, Cho SJ, Stout-Delgado H. Harris R, et al. Int J Mol Sci. 2021 Nov 9;22(22):12097. doi: 10.3390/ijms222212097. Int J Mol Sci. 2021. PMID: 34829979 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous