Predicting response to checkpoint inhibitors in melanoma beyond PD-L1 and mutational burden - PubMed (original) (raw)
doi: 10.1186/s40425-018-0344-8.
Carl Morrison 1 2 3 4, Jeffrey M Conroy 6 5, Mary K Nesline 5, Sean T Glenn 7 5, Devin Dressman 5, Antonios Papanicolau-Sengos 5, Blake Burgher 5, Jonathan Andreas 5, Vincent Giamo 5, Moachun Qin 5, Yirong Wang 5, Felicia L Lenzo 5, Angela Omilian 8, Wiam Bshara 8, Matthew Zibelman 9, Pooja Ghatalia 9, Konstantin Dragnev 10, Keisuke Shirai 10, Katherine G Madden 10, Laura J Tafe 10 11, Neel Shah 12, Deepa Kasuganti 12, Luis de la Cruz-Merino 13, Isabel Araujo 13, Yvonne Saenger 14, Margaret Bogardus 14, Miguel Villalona-Calero 15, Zuanel Diaz 15, Roger Day 16, Marcia Eisenberg 17, Steven M Anderson 17, Igor Puzanov 18, Lorenzo Galluzzi 19 20 21, Mark Gardner 5, Marc S Ernstoff 18
Affiliations
- PMID: 29743104
- PMCID: PMC5944039
- DOI: 10.1186/s40425-018-0344-8
Predicting response to checkpoint inhibitors in melanoma beyond PD-L1 and mutational burden
Carl Morrison et al. J Immunother Cancer. 2018.
Abstract
Background: Immune checkpoint inhibitors (ICIs) have changed the clinical management of melanoma. However, not all patients respond, and current biomarkers including PD-L1 and mutational burden show incomplete predictive performance. The clinical validity and utility of complex biomarkers have not been studied in melanoma.
Methods: Cutaneous metastatic melanoma patients at eight institutions were evaluated for PD-L1 expression, CD8+ T-cell infiltration pattern, mutational burden, and 394 immune transcript expression. PD-L1 IHC and mutational burden were assessed for association with overall survival (OS) in 94 patients treated prior to ICI approval by the FDA (historical-controls), and in 137 patients treated with ICIs. Unsupervised analysis revealed distinct immune-clusters with separate response rates. This comprehensive immune profiling data were then integrated to generate a continuous Response Score (RS) based upon response criteria (RECIST v.1.1). RS was developed using a single institution training cohort (n = 48) and subsequently tested in a separate eight institution validation cohort (n = 29) to mimic a real-world clinical scenario.
Results: PD-L1 positivity ≥1% correlated with response and OS in ICI-treated patients, but demonstrated limited predictive performance. High mutational burden was associated with response in ICI-treated patients, but not with OS. Comprehensive immune profiling using RS demonstrated higher sensitivity (72.2%) compared to PD-L1 IHC (34.25%) and tumor mutational burden (32.5%), but with similar specificity.
Conclusions: In this study, the response score derived from comprehensive immune profiling in a limited melanoma cohort showed improved predictive performance as compared to PD-L1 IHC and tumor mutational burden.
Keywords: Algorithmic analysis; Borderline; Immune Desert; Inflamed; Ipilimumab; Nivolumab; Pembrolizumab.
Conflict of interest statement
Ethics approval and consent to participate
Existing deidentified specimens and associated clinical data was submitted to OmniSeq from Roswell Park Comprehensive Cancer Center (Buffalo, NY), Fox Chase Cancer Center (Philadelphia, PA), Dartmouth-Hitchcock Medical Center (Lebanon, NH), Northwest Oncology (Munster, IN), Hospital Universitario Virgen Macarena (Sevilla, Spain), Columbia University (New York, NY), and Miami Cancer Institute (Miami, FL) with institutional review board approval from each individual institution. OmniSeq’s analysis utilized deidentified data that was considered non-human subjects research under IRB-approved protocol (BDR #073166) at Roswell Park Comprehensive Cancer Center (Buffalo, NY).
Competing interests
Disclosures: MZ, PG, KD, KS, KGM, LJT, NS, DK, LDLCM, IA, YS, MB, MVC, and ZD have no conflicts of interest to disclose. SP, JMC, MKN, STG, DD, APS, BB, JA, VG, MQ, YW, FLL, MG, and CM are all employees of OmniSeq, Inc. (Buffalo, NY) and hold restricted stock in OmniSeq, Inc. AO, WB, IP, CM, STG, JMC, and MSE are employees of Roswell Park Comprehensive Cancer Center (Buffalo, NY). Roswell Park Comprehensive Cancer Center is the majority shareholder of OmniSeq, Inc. MSE, RD, and LG provide remunerated consulting to OmniSeq, Inc. ME and SMA are employees of Laboratory Corporation of America Holdings (Burlington, NC). Laboratory Corporation of America Holdings, (Burlington, NC) is a shareholder in OmniSeq, Inc.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
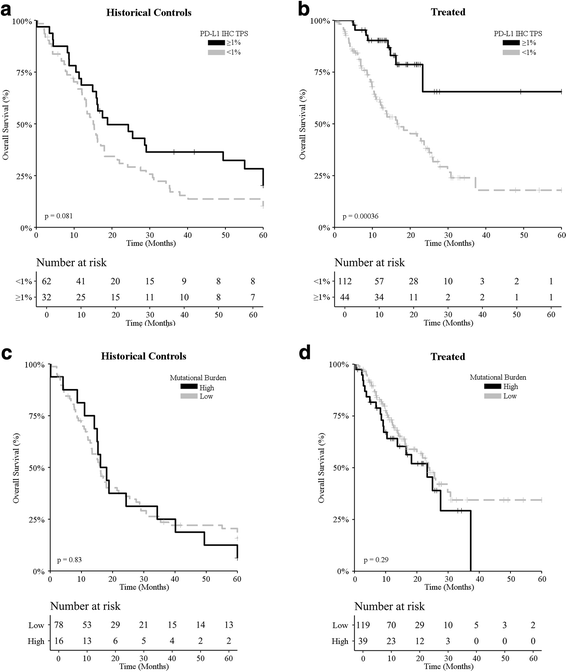
Survival analysis of melanoma patients based on PD-L1 expression levels and mutational burden. a, b Overall survival upon stratification based on PD-L1 positivity (tumor proportion score ≥ 1% versus < 1%) for metastatic melanoma patients treated (a) prior to the introduction of ICIs (historical controls; n = 94) and (b) ICI-treated melanoma patients (n = 137). c, d Overall survival upon stratification based on mutational burden [high (≥ 7.1 mut/Mb) versus non-high] for metastatic melanoma patients treated (c) prior to the introduction of ICIs (historical controls; n = 94) and (d) ICI-treated melanoma patients (n = 137). _p_-values are indicated
Fig. 2
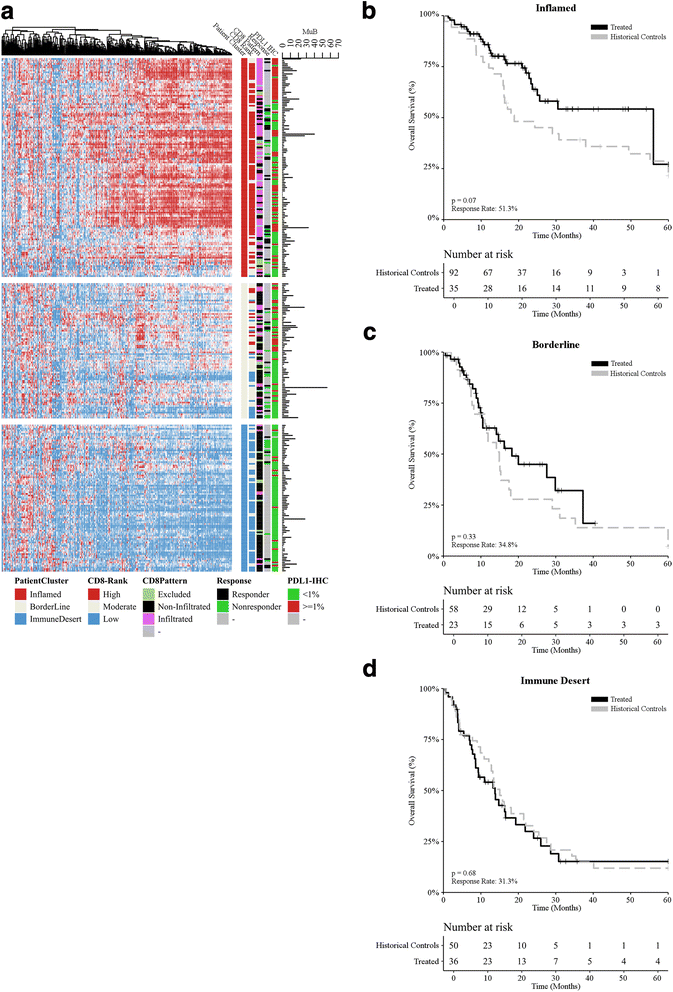
Immunological landscape of melanoma and its association with multiple variables. a Unsupervised hierarchical clustering (rows = patients, columns = genes) for 394 immune transcripts identified three major groups defined as “inflamed” (group 1), “borderline” (group 2), and “immune desert” (group 3). CD8 (assessed by RNA-seq) and PD-L1 expression levels (assessed by immunohistochemistry), response to ICIs (as per RECIST v.1.1), CD8 infiltration pattern (assessed by a trained pathologist, CM and APS), and mutational burden (assessed by whole-exon sequencing) are depicted. b-d Overall survival for melanoma patients from the (b) inflamed (n = 131), (c) borderline (n = 81), and (d) immune desert (n = 88) groups upon stratification based on treatment (historical controls versus ICI-based immunotherapy). p values are reported
Fig. 3
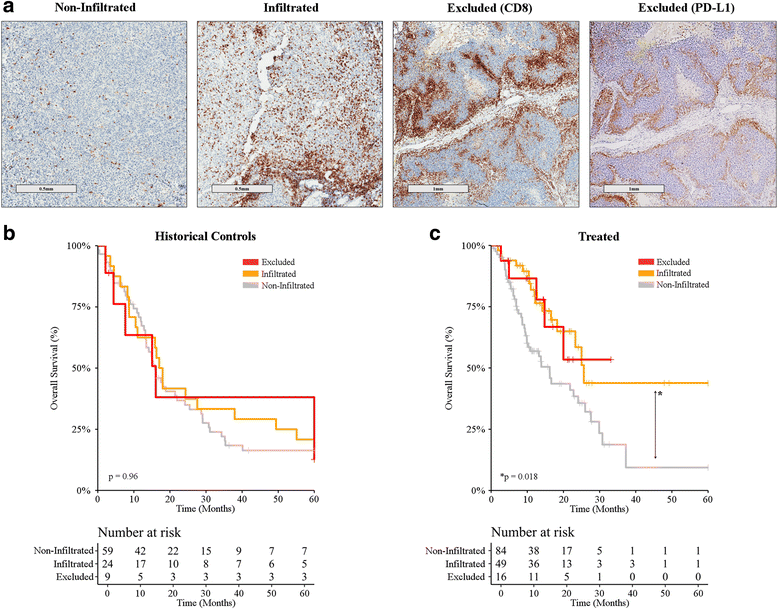
CD8+ T-cell infiltration pattern and clinical benefits from immune checkpoint inhibition. a CD8+ T-cell infiltration pattern was assessed by a trained pathologist upon immunohistochemistry with a CD8-specific antibody. Representative images are depicted (scale bar = 500 μm or 1 mm). b, c Overall survival upon stratification based on infiltration pattern (non-infiltrated, infiltrated, excluded) for metastatic melanoma patients treated (b) prior to the introduction of ICIs (historical controls; n = 94) and (c) ICI-treated melanoma patients (n = 137). For all comparisons p > 0.05
Fig. 4
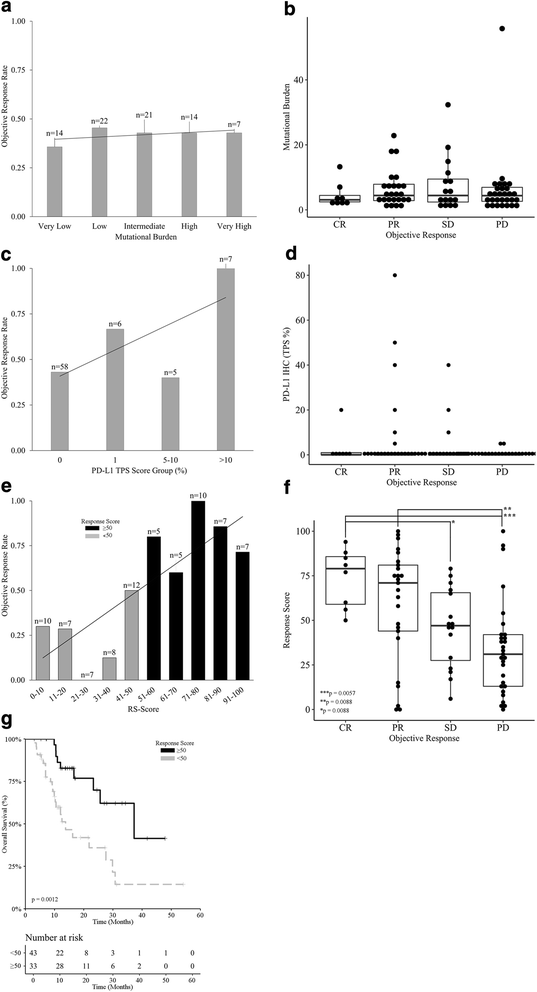
Predictive performance of the RS as compared to PD-L1 expression levels and mutational burden. a Response rates for mutational burden using a scale of very low, low, intermediate, high, and very high. While increasing response rates occur with an increase in mutational burden the range of values from 36% to 45% is also limited for clinical use. b Box plot shows a pair-wise comparison of mutational burden for CR, PR, SD, and PD. c Response rates for PD-L1 IHC using a tumor proportion score (TPS) with values of zero or negative, 1–4%, 5–10%, and > 10%. While increasing response rates occur with an increase in TPS the range of values from 40% to 100% is limited for clinical use. d Box plot shows a pair-wise comparison of PD-L1 IHC TPS for CR, PR, SD, and PD. e Objective response rates in groups of ICI-treated melanoma patients stratified by RS. Linear regression supports a dynamic range from 30% to 100%. f Box plot shows a pair-wise comparison of RS values for CR, PR, SD, and PD. g Overall survival upon stratification based on RS (≥ 50 versus < 50) for ICI-treated melanoma patients (n = 137), p value is indicated, for comprehensive immune profiling using response score (RS) with a bin width of 10. Response rate shows a dynamic range of values from near zero to greater than 95% with increasing RS. Survival curve for patients with a RS ≥ 50 and < 50 shows an improved survival for the former (p = 0.0012)
Similar articles
- Value of PD-L1, PD-1, and CTLA-4 Expression in the Clinical Practice as Predictors of Response to Nivolumab and Ipilimumab in Monotherapy in Patients With Advanced Stage Melanoma.
Santos-Briz A, Cañueto J, Carmen SD, Barrios B, Yuste M, Bellido L, Ludeña MD, Román C. Santos-Briz A, et al. Am J Dermatopathol. 2021 Jun 1;43(6):423-428. doi: 10.1097/DAD.0000000000001856. Am J Dermatopathol. 2021. PMID: 33395045 - The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma.
Mahoney KM, Freeman GJ, McDermott DF. Mahoney KM, et al. Clin Ther. 2015 Apr 1;37(4):764-82. doi: 10.1016/j.clinthera.2015.02.018. Epub 2015 Mar 29. Clin Ther. 2015. PMID: 25823918 Free PMC article. Review. - Association Between Sex and Immune Checkpoint Inhibitor Outcomes for Patients With Melanoma.
Jang SR, Nikita N, Banks J, Keith SW, Johnson JM, Wilson M, Lu-Yao G. Jang SR, et al. JAMA Netw Open. 2021 Dec 1;4(12):e2136823. doi: 10.1001/jamanetworkopen.2021.36823. JAMA Netw Open. 2021. PMID: 34854905 Free PMC article. - A PD-1/PD-L1 Proximity Assay as a Theranostic Marker for PD-1 Blockade in Patients with Metastatic Melanoma.
Girault I, Adam J, Shen S, Roy S, Brard C, Faouzi S, Routier E, Lupu J, Warren S, Sorg K, Ong S, Morel P, Scoazec JY, Vagner S, Robert C. Girault I, et al. Clin Cancer Res. 2022 Feb 1;28(3):518-525. doi: 10.1158/1078-0432.CCR-21-1229. Epub 2021 Nov 16. Clin Cancer Res. 2022. PMID: 34785583 - Current landscape and future of dual anti-CTLA4 and PD-1/PD-L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (NSCLC).
Chae YK, Arya A, Iams W, Cruz MR, Chandra S, Choi J, Giles F. Chae YK, et al. J Immunother Cancer. 2018 May 16;6(1):39. doi: 10.1186/s40425-018-0349-3. J Immunother Cancer. 2018. PMID: 29769148 Free PMC article. Review.
Cited by
- Immune Checkpoint Inhibitor Therapy for Metastatic Melanoma: What Should We Focus on to Improve the Clinical Outcomes?
Hossain SM, Ly K, Sung YJ, Braithwaite A, Li K. Hossain SM, et al. Int J Mol Sci. 2024 Sep 20;25(18):10120. doi: 10.3390/ijms251810120. Int J Mol Sci. 2024. PMID: 39337605 Free PMC article. Review. - A Retrospective Analysis of the Prognostic Factors and Adverse Events in the Treatment of Mucosal Melanoma in a Single Centre.
Wesener L, Hagelstein V, Terheyden P, Langan EA. Wesener L, et al. J Clin Med. 2024 Aug 13;13(16):4741. doi: 10.3390/jcm13164741. J Clin Med. 2024. PMID: 39200883 Free PMC article. - Populations of triple negative and hormone receptor positive HER2 negative breast tumors share immune gene profiles.
Stanton S, Schmitz F, Copeland W, DellAringa J, Newhall K, Disis M. Stanton S, et al. Res Sq [Preprint]. 2024 Aug 2:rs.3.rs-4542494. doi: 10.21203/rs.3.rs-4542494/v1. Res Sq. 2024. PMID: 39149486 Free PMC article. Preprint. - Predicting immune checkpoint therapy response in three independent metastatic melanoma cohorts.
Szadai L, Bartha A, Parada IP, Lakatos AIT, Pál DMP, Lengyel AS, de Almeida NP, Jánosi ÁJ, Nogueira F, Szeitz B, Doma V, Woldmar N, Guedes J, Ujfaludi Z, Pahi ZG, Pankotai T, Kim Y, Győrffy B, Baldetorp B, Welinder C, Szasz AM, Betancourt L, Gil J, Appelqvist R, Kwon HJ, Kárpáti S, Kuras M, Murillo JR, Németh IB, Malm J, Fenyö D, Pawłowski K, Horvatovich P, Wieslander E, Kemény LV, Domont G, Marko-Varga G, Sanchez A. Szadai L, et al. Front Oncol. 2024 Jul 2;14:1428182. doi: 10.3389/fonc.2024.1428182. eCollection 2024. Front Oncol. 2024. PMID: 39015503 Free PMC article. - AUNP-12 Near-Infrared Fluorescence Probes across NIR-I to NIR-II Enable In Vivo Detection of PD-1/PD-L1 Axis in the Tumor Microenvironment.
Zhang X, Wang P, Shi G, Tang C, Xue H. Zhang X, et al. Bioconjug Chem. 2024 Jul 17;35(7):1064-1074. doi: 10.1021/acs.bioconjchem.4c00266. Epub 2024 Jul 9. Bioconjug Chem. 2024. PMID: 38980173 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials