ISG15 in antiviral immunity and beyond - PubMed (original) (raw)
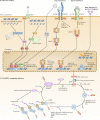
Fig. 1. The ISG15 conjugation pathway.
a | Induction of ubiquitin-like protein ISG15. The expression of ubiquitin-like protein ISG15 and members of the ISG15 conjugation pathway is strongly induced by type I interferons (IFNs). Upon type I interferon stimulation, interferon regulatory factor 9 (IRF9) interacts with phosphorylated signal transducer and activator of transcription 1 (STAT1) and STAT2 and forms the interferon-stimulated gene factor 3 (ISGF3) complex, which recognizes the interferon-sensitive response element (ISRE) promoters of ISG15 and its conjugation enzymes to induce their expression. During chronic viral infection, both IFNβ and type III interferons lead to the formation of an unphosphorylatyed ISGF3 (U-ISGF3) complex, which is composed of IRF9 and unphosphorylated STAT1–STAT2 (refs,), to induce the expression of a subset of interferon-stimulated genes (ISGs), including ISG15 (ref.). Additional stimuli, including lipopolysaccharide (LPS), foreign DNA or RNA, retinoic acid and DNA-damaging agents, are also able to induce the expression of ISG15. LPS was shown to induce ISG15 expression via TIR domain-containing adapter protein inducing IFNβ (TRIF; also known as TICAM1)–IRF3 signalling pathways, but this stimulation still functioned in a type I interferon-dependent manner. Retinoic acid can also induce ISG15 expression through its induction of type I interferons,. By contrast, the introduction of double-stranded RNA (dsRNA) could induce ISG15 expression via IRF3 in an interferon-independent manner. IRF3 forms a complex with CREB-binding protein (CREBBP)/p300 and binds to the ISRE elements to induce ISG15 expression,. Bacterial DNA can also induce ISG15 expression independent of type I interferon and can signal through a cytosolic surveillance pathway that requires stimulator of interferon genes protein (STING), serine/threonine-protein kinase TBK1, IRF3 and IRF7 (ref.). Notably, the expression of ISG15 and its conjugation enzymes is controlled by the ISRE elements, mediated by various IRFs. However, a recent study found that ISG15, as well as its conjugation enzymes, including ubiquitin-activating enzyme E1 homologue (UBE1L), ubiquitin/ISG15-conjugating enzyme E2 L6 (UBCH8) and E3 ubiquitin/ISG15 ligase TRIM25, possess cellular tumour antigen p53-responsive elements in their promoters. DNA-damaging (for example, ultraviolet light or doxorubicin) or agents (for example, camptothecin) induce p53, which binds to these p53-responsive elements to transactivate the ISG15 pathway. b | The ISG15 conjugation pathway. ISG15 is initially expressed as a 17 kDa precursor protein that is then processed into its mature 15 kDa form via protease cleavage,. Conjugation of ISG15, commonly referred to as ISGylation, utilizes a three-step enzymatic cascade similar to that of ubiquitination. The activating E1 enzyme UBE1L functions only in the ISG15 pathway,, forming an ATP-dependent thioester bond with ISG15 (ref.). Following activation, ISG15 is transferred to the active-site cysteine of the ubiquitin/ISG15-conjugating enzyme E2 L6 (UBCM8) (in mouse) and UBCH8 (in human),. Finally, an E3 ligase facilitates the conjugation of ISG15 to target proteins. Although several enzymes, including E3 ubiquitin-protein ligase ARIH1 and TRIM25, have been found to act as E3 ligases within the ISG15 pathway,, human HERC5 (mouse HERC6) appears to be the dominant E3 ligase that coordinates the conjugation of the majority of substrates to ISG15 (refs,–). However, during DNA-damage responses (DDRs),TRIM25 also serves as a p53-specific ISG15 E3 ligase. Notably, its corresponding E2 enzyme for ISG15 conjugation has not been identified. HERC5 associates with polyribosomes and appears to target newly synthesized proteins, including viral proteins and many interferon-induced proteins. ISGylation is also reversible via the specific removal of ISG15 from conjugated proteins by the deconjugating enzyme Ubl carboxy-terminal hydrolase 18 (USP18),. cGAS, cyclic GMP-AMP synthase; ER, endoplasmic reticulum; HCV, hepatitis C virus; IFNAR1, interferon α/β receptor 1; IFNAR2, interferon α/β receptor 2; IFNLR1, interferon-λ receptor 1; IL-10R2, interleukin-10 receptor subunit 2; JAK1, Janus kinase 1; MAVS, mitochondrial antiviral-signalling protein; RIG-I, retinoic acid-inducible gene I; ssDNA, single-stranded DNA; TLR4, Toll-like receptor 4.
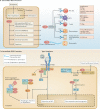
Fig. 2. Extracellular and intracellular functions of unconjugated ISG15.
a | Extracellular functions. Several immunomodulatory and cytokine functions have been ascribed to ubiquitin-like protein ISG15. Although it does not possess a leader signal sequence to direct its secretion, ISG15 has been found to be released through microparticles, exosomes and neutrophilic granules; from secretory lysosomes; and via apoptosis. It has been described as influencing the functions of several different types of cells. Natural killer (NK) cells: ISG15 induces NK cell proliferation and stimulates interferon-γ (IFNγ) production. Dendritic cells (DCs): ISG15 induces DC maturation and upregulates the expression of E-cadherin, CD15 and CD86 (ref.). Neutrophils: ISG15 functions as a chemotactic factor and an activator of neutrophils. T cells: ISG15 was found to induce the secretion of IFNγ. Macrophages: ISG15-containing microparticles and exosomes have been found to stimulate macrophages to secrete pro-inflammatory cytokines and mediate anti-HIV activity, respectively. It remains to be determined whether it is a stand-alone effect of ISG15 or an effect synergistic with those of other molecules in the secretory vesicles. Lung epithelium cells: recombinant ISG15 reduces CXC-chemokine ligand 10 (CXCL10) protein production in human rhinovirus-16 (HRV-16)-infected human bronchial epithelial (HBE) cells. ISG15 receptor: lymphocyte function-associated antigen 1 (LFA1; also known as αLβ2 integrin, or CD11a/CD18) was recently identified as a cell surface receptor for extracellular ISG15. Binding of ISG15 to LFA1 on NK cells stimulated the release of IFNγ and interleukin-10 (IL-10) from IL-12 primed NK cells. Whether LFA1 senses extracellular ISG15 in other cell types remains to be determined. b | Intracellular functions: Ubl carboxy-terminal hydrolase 18 (USP18) and S-phase kinase-associated protein 2 (SKP2): USP18, which is induced by type I interferons, mediates the negative feedback regulation of interferon signalling independent of its deISGylase activity. USP18 is recruited by signal transducer and activator of transcription (STAT2) and binds to interferon α/β receptor 2 (IFNAR2) of the interferon receptor complex, displacing Janus kinase 1 (JAK1) and suppressing interferon signalling and the downstream expression of interferon-stimulated genes (ISGs). Notably, human ISG15 binds to USP18 and inhibits SKP2-mediated ubiquitylation and proteasomal-mediated degradation,. In the absence of human ISG15, human USP18 is degraded, and this allows for continued JAK1 binding to the IFNAR2 complex and prolonged IFNAR signalling and ISG expression. By contrast, mouse ISG15 does not alter the stability of mouse USP18 and its ability to suppress IFNAR signalling, although the precise reason for this difference is still unclear, as discussed in Box 1 (ref.). Histone deacetylase 6 (HDAC6) and ubiquitin-binding protein p62 (also known as SQSTM1): intracellular ISG15 binds to HDAC6 and p62 to regulate autophagic clearance of proteins, especially ISGylated proteins. Leucine-rich repeat-containing protein 25 (LRRC25)–retinoic acid-inducible gene I protein (RIG-I)–p62: intracellular ISG15 binds to LRRC25, p62 and RIG-I to mediate the autophagic degradation of RIG-I, which is critical to LRRC25-mediated downregulation of type I interferon signalling. Hypoxia inducible factor 1α (HIF1α): ISG15 has been shown to interact with HIF1α, preventing its dimerization with HIF1β to initiate downstream gene expression. E3 ubiquitin-protein ligase NEDD4: ISG15 has been shown to bind to the NEDD4 and disrupt its interaction with ubiquitin E2 conjugating enzymes, preventing ubiquitin from being transferred to NEDD4 for ubiquitylation. The E3 ubiquitin ligase activity of NEDD4 is critical to Ebola virus-like particle virus budding,. HBMECs, human brain microvascular endothelial cells; MACs, macrophages.
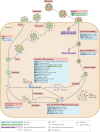
Fig. 3. Direct effects of ISG15 on viral replication.
In general, the viral replication cycle consists of the following steps: attachment, entry, uncoating, viral replication complex formation, transcription, translation, post-translational modification and processing of viral proteins, packaging, capsid assembly, egress and trafficking, assembly and budding, release and virion maturation. Ubiquitin-like protein ISG15 was found to restrict the infection of murine norovirus at the virus entry or uncoating step. This inhibition was conjugation-dependent, but the precise mechanism is still unknown. Viral DNA or RNA replication: many studies of the antiviral activity of ISG15 analysed the replication of viral DNA or RNA in the presence or absence of ISG15 and its conjugation machinery. During IBV infection, ISG15 disrupts the viral replication machinery through conjugation to the viral nucleoprotein (NP), disrupting its ability to oligomerize and form viral nucleoproteins (vRNPs), thus inhibiting viral RNA synthesis. In addition, the replication of many viruses (blue) was inhibited by ISG15 (refs,,,,,,,,–), yet the precise mechanisms by which ISG15 inhibits their replication remain unclear. During chronic hepatitis C virus (HCV) infection (green), ISG15 appeared to have a proviral role, because its overexpression reduced the responsiveness of the cell to interferon-α (IFNα) by maintaining the abundance of Ubl carboxy-terminal hydrolase 18 (USP18), a negative regulator of type I interferon receptor signalling. In post-translational modification and processing, ISG15 conjugation has been shown to target newly synthesized proteins, including viral proteins and several interferon-stimulated genes during the type I interferon response. ISGylation of viral proteins acts as a host antiviral strategy. ISGylation of such proteins elicits antiviral effects by impairing their function, for example, essential effectors of viral replication complex (influenza B virus (IBV) NP and influenza A virus (IAV) NS1/A), proteins involved in host shut-off (coxsackie virus 3 (CVB3) 2A protease (2APro)), counteractors of host immunity (human cytomegalovirus (HCMV) pUL26 (ref.)) and capsid proteins (human papilloma virus (HPV)-16 L1). Egress and trafficking and assembly and budding: both ISG15 and ISG15 conjugation have been shown to restrict intracellular trafficking of viruses and/or subsequent viral release from the cell surface,, mostly by targeting host proteins necessary for trafficking (for example, tumour susceptibility gene 101 protein (TSG101)) or for release and/or budding (for example, ubiquitin protein ligase NEDD4 (refs,)) and preventing their interactions with either viral proteins or members of the host transportation complex,. Viral release: for specific viruses, ISG15 was found to modulate the amount of released infectious viral particles while intracellular viral replication remains intact. Interestingly, whereas the molecular mechanism is unclear, such effects can be either antiviral (for example, dengue virus 2 (DENV-2)) or proviral (for example, hepatitis B virus (HBV)) depending on the type of virus. Viral latency: ISG15 was also shown to maintain viral latency and prevent reactivation of Kaposi's sarcoma-associated herpesvirus (KSHV) infection via the regulation of specific KSHV microRNAs,. ASLV, avian sarcoma leukosis virus; CHMP5, charged multivesicular body protein 5; CVB3, coxsackievirus B3; EBOV, Ebola virus; HA, haemagglutinin; MHV-3, murine hepatitis virus 3; MNV-1, murine norovirus 1; NDV, Newcastle disease virus; NS1, non-structural protein 1; RSV, respiratory syncytial virus; SeV, Sendai virus; WNV, West Nile virus; ZIKV, Zika virus.
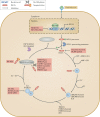
Fig. 4. Viral immune-evasion strategies targeting the ISG15 pathway.
Viral immune-evasion strategies have been identified that target the ubiquitin-like protein ISG15 pathway. Certain viruses, for example, human cytomegalovirus (HCMV), and hepatitis C virus (HCV),, suppress the expression of ISG15 at the transcriptional level. Vaccinia virus E3L and HCMV pUL26 (ref.) proteins bind to ISG15 to inhibit ISG15 conjugation by undefined mechanisms. HCMV pUL26 (ref.), influenza A virus (IAV) non-structural protein 1 (NS1) and Kaposi's sarcoma-associated herpesvirus (KSHV) viral interferon regulatory factor 1 (vIRF1) also interact with an E3 ISG15–protein ligase HERC5 to reduce protein conjugation by ISG15. Even when the viral and host proteins are conjugated to ISG15, viruses have also developed strategies to deconjugate or sequester ISGylated viral proteins. Influenza B virus (IBV) NS1 protein counteracts ISG15 antiviral activity by sequestering ISGylated viral proteins, particularly viral nucleoproteins (NPs), to prevent their incorporation into NP oligomers, thus disrupting viral RNA synthesis. Several virus families encode enzymes possessing deISGylase activities, including papain-like protease (PLpro) of coronaviruses and viral ovarian tumour domain (OTU) proteases of nairoviruses and arteriviruses, to remove ISG15 from conjugated proteins. CCHFV, Crimean–Congo haemorrhagic fever virus; DUGV, Dugbe virus; EAV, equine arteritis virus; HERC5, E3 ISG15–protein ligase HERC5; HERC6, E3 ISG15–protein ligase HERC6; IE1, immediate-early protein 1; ISRE, interferon-stimulated response elements; MERS, Middle East respiratory syndrome; NS3, non-structural protein 3; PRRSV, porcine respiratory and reproductive syndrome virus; RNAP II, RNA polymerase II; SARS, severe acute respiratory syndrome; TRIM25, E3 ubiquitin/ISG15 ligase; UBCH8, ubiquitin/ISG15-conjugating enzyme E2 L6; UBCM8, ubiquitin/ISG15-conjugating enzyme E2 L6; UBE1L, ubiquitin-activating enzyme E1 homologue; USP18, Ubl carboxy-terminal hydrolase 18.