ComplexContact: a web server for inter-protein contact prediction using deep learning - PubMed (original) (raw)
ComplexContact: a web server for inter-protein contact prediction using deep learning
Hong Zeng et al. Nucleic Acids Res. 2018.
Abstract
ComplexContact (http://raptorx2.uchicago.edu/ComplexContact/) is a web server for sequence-based interfacial residue-residue contact prediction of a putative protein complex. Interfacial residue-residue contacts are critical for understanding how proteins form complex and interact at residue level. When receiving a pair of protein sequences, ComplexContact first searches for their sequence homologs and builds two paired multiple sequence alignments (MSA), then it applies co-evolution analysis and a CASP-winning deep learning (DL) method to predict interfacial contacts from paired MSAs and visualizes the prediction as an image. The DL method was originally developed for intra-protein contact prediction and performed the best in CASP12. Our large-scale experimental test further shows that ComplexContact greatly outperforms pure co-evolution methods for inter-protein contact prediction, regardless of the species.
Figures
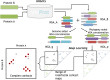
Figure 1.
Illustration of ComplexContact workflow. Given a pair of putative interacting proteins A and B, ComplexContact first uses HHblits (38) to search for sequence homologs and build an MSA for each protein. Then ComplexContact constructs two paired MSAs using genome and phylogeny information. Finally, ComplexContact applies deep learning to predict two inter-protein contact maps from the two paired MSAs and calculates their average as the final contact prediction. The top half of this figure is inspired by Fig. S2 in (40).
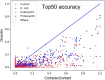
Figure 2.
The top-50 prediction accuracy by ComplexContact and Gremlin on the protein pairs extracted from 3DComplex. Each dot represents one protein pair and is colored by its species. A dot below the diagonal line indicates that ComplexContact has a better accuracy.
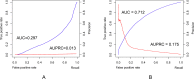
Figure 3.
Quality assessment of the top 50 predicted interfacial contacts for 4479 heterodimers extracted from 3Dcomplex. (A) and (B) show the precision-recall (in red) and ROC (in blue) curves generated by Gremlin and ComplexContact, respectively. AUC: Area Under the ROC curve; AUPRC: Area Under the precision-recall curve.
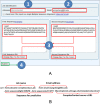
Figure 4.
ComplexContact server job submission. (A) Users may submit a job by a web interface, which has fields for job name (1), optional user email address (2), and a pair of sequences (or multiple sequence alignments) (3). The sequences shall be in FASTA format and can also be submitted in a file. (B) Users may also submit a job by a publicly available program Curl without using the web interface. In this command, Job name and Email address are optional. A job URL will be returned on screen after submission. Curl allows users to submit a large number of jobs quickly.
Figure 5.
ComplexContact server result page. The left part shows the predicted complex contact map (1), where the predicted probability is displayed in greyscale, with a darker color indicating a larger value. The middle part shows three panels. The first one is used to zoom and drag contact images (2). The second panel is for downloading the predicted contact map (3), and the third panel is for downloading the detailed prediction results (4). The right part shows two paired MSAs generated by genome-based method (5) and phylogeny-based method (6).
Similar articles
- A Web-Based Protocol for Interprotein Contact Prediction by Deep Learning.
Jing X, Zeng H, Wang S, Xu J. Jing X, et al. Methods Mol Biol. 2020;2074:67-80. doi: 10.1007/978-1-4939-9873-9_6. Methods Mol Biol. 2020. PMID: 31583631 - Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model.
Wang S, Sun S, Li Z, Zhang R, Xu J. Wang S, et al. PLoS Comput Biol. 2017 Jan 5;13(1):e1005324. doi: 10.1371/journal.pcbi.1005324. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28056090 Free PMC article. - Analysis of deep learning methods for blind protein contact prediction in CASP12.
Wang S, Sun S, Xu J. Wang S, et al. Proteins. 2018 Mar;86 Suppl 1(Suppl 1):67-77. doi: 10.1002/prot.25377. Epub 2017 Sep 6. Proteins. 2018. PMID: 28845538 Free PMC article. - The trRosetta server for fast and accurate protein structure prediction.
Du Z, Su H, Wang W, Ye L, Wei H, Peng Z, Anishchenko I, Baker D, Yang J. Du Z, et al. Nat Protoc. 2021 Dec;16(12):5634-5651. doi: 10.1038/s41596-021-00628-9. Epub 2021 Nov 10. Nat Protoc. 2021. PMID: 34759384 Review. - Recent Applications of Deep Learning Methods on Evolution- and Contact-Based Protein Structure Prediction.
Suh D, Lee JW, Choi S, Lee Y. Suh D, et al. Int J Mol Sci. 2021 Jun 2;22(11):6032. doi: 10.3390/ijms22116032. Int J Mol Sci. 2021. PMID: 34199677 Free PMC article. Review.
Cited by
- Deep learning methods for 3D structural proteome and interactome modeling.
Lee D, Xiong D, Wierbowski S, Li L, Liang S, Yu H. Lee D, et al. Curr Opin Struct Biol. 2022 Apr;73:102329. doi: 10.1016/j.sbi.2022.102329. Epub 2022 Feb 6. Curr Opin Struct Biol. 2022. PMID: 35139457 Free PMC article. Review. - Deep graph learning of inter-protein contacts.
Xie Z, Xu J. Xie Z, et al. Bioinformatics. 2022 Jan 27;38(4):947-953. doi: 10.1093/bioinformatics/btab761. Bioinformatics. 2022. PMID: 34755837 Free PMC article. - Inter-protein residue covariation information unravels physically interacting protein dimers.
Salmanian S, Pezeshk H, Sadeghi M. Salmanian S, et al. BMC Bioinformatics. 2020 Dec 17;21(1):584. doi: 10.1186/s12859-020-03930-7. BMC Bioinformatics. 2020. PMID: 33334319 Free PMC article. - Genetic interaction mapping informs integrative structure determination of protein complexes.
Braberg H, Echeverria I, Bohn S, Cimermancic P, Shiver A, Alexander R, Xu J, Shales M, Dronamraju R, Jiang S, Dwivedi G, Bogdanoff D, Chaung KK, Hüttenhain R, Wang S, Mavor D, Pellarin R, Schneidman D, Bader JS, Fraser JS, Morris J, Haber JE, Strahl BD, Gross CA, Dai J, Boeke JD, Sali A, Krogan NJ. Braberg H, et al. Science. 2020 Dec 11;370(6522):eaaz4910. doi: 10.1126/science.aaz4910. Science. 2020. PMID: 33303586 Free PMC article. - InterEvDock3: a combined template-based and free docking server with increased performance through explicit modeling of complex homologs and integration of covariation-based contact maps.
Quignot C, Postic G, Bret H, Rey J, Granger P, Murail S, Chacón P, Andreani J, Tufféry P, Guerois R. Quignot C, et al. Nucleic Acids Res. 2021 Jul 2;49(W1):W277-W284. doi: 10.1093/nar/gkab358. Nucleic Acids Res. 2021. PMID: 33978743 Free PMC article.
References
- Lensink M.F., Velankar S., Wodak S.J.. Modeling protein–protein and protein–peptide complexes: CAPRI 6th edition. Proteins: Struct. Funct. Bioinform. 2017; 85:359–377. - PubMed
- Mosca R., Céol A., Aloy P.. Interactome3D: adding structural details to protein networks. Nat. Methods. 2013; 10:47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources