Leveraging Existing 16S rRNA Gene Surveys To Identify Reproducible Biomarkers in Individuals with Colorectal Tumors - PubMed (original) (raw)
Meta-Analysis
Leveraging Existing 16S rRNA Gene Surveys To Identify Reproducible Biomarkers in Individuals with Colorectal Tumors
Marc A Sze et al. mBio. 2018.
Erratum in
- Erratum for Sze and Schloss, "Leveraging Existing 16S rRNA Gene Surveys To Identify Reproducible Biomarkers in Individuals with Colorectal Tumors".
Sze MA, Schloss PD. Sze MA, et al. mBio. 2018 Oct 16;9(5):e02076-18. doi: 10.1128/mBio.02076-18. mBio. 2018. PMID: 30327440 Free PMC article. No abstract available.
Abstract
An increasing body of literature suggests that both individual and collections of bacteria are associated with the progression of colorectal cancer. As the number of studies investigating these associations increases and the number of subjects in each study increases, a meta-analysis to identify the associations that are the most predictive of disease progression is warranted. We analyzed previously published 16S rRNA gene sequencing data collected from feces and colon tissue. We quantified the odds ratios (ORs) for individual bacterial taxa that were associated with an individual having tumors relative to a normal colon. Among the fecal samples, there were no taxa that had significant ORs associated with adenoma and there were 8 taxa with significant ORs associated with carcinoma. Similarly, among the tissue samples, there were no taxa that had a significant OR associated with adenoma and there were 3 taxa with significant ORs associated with carcinoma. Among the significant ORs, the association between individual taxa and tumor diagnosis was equal to or below 7.11. Because individual taxa had limited association with tumor diagnosis, we trained Random Forest classification models using only the taxa that had significant ORs, using the entire collection of taxa found in each study, and using operational taxonomic units defined based on a 97% similarity threshold. All training approaches yielded similar classification success as measured using the area under the curve. The ability to correctly classify individuals with adenomas was poor, and the ability to classify individuals with carcinomas was considerably better using sequences from feces or tissue.IMPORTANCE Colorectal cancer is a significant and growing health problem in which animal models and epidemiological data suggest that the colonic microbiota have a role in tumorigenesis. These observations indicate that the colonic microbiota is a reservoir of biomarkers that may improve our ability to detect colonic tumors using noninvasive approaches. This meta-analysis identifies and validates a set of 8 bacterial taxa that can be used within a Random Forest modeling framework to differentiate individuals as having normal colons or carcinomas. When models trained using one data set were tested on other data sets, the models performed well. These results lend support to the use of fecal biomarkers for the detection of tumors. Furthermore, these biomarkers are plausible candidates for further mechanistic studies into the role of the gut microbiota in tumorigenesis.
Keywords: 16S rRNA; adenoma; biomarkers; carcinoma; colorectal cancer; diagnostic; feces; microbiome.
Copyright © 2018 Sze and Schloss.
Figures
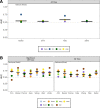
FIG 1
Comparison of alpha diversity indices that were significant between individuals with normal colons and those with adenomas or carcinomas using data collected from fecal samples. (A) Comparison of evenness between individuals with normal colons and adenomas. (B) Comparison of evenness between individuals with normal colons and carcinomas. (C) Comparison of Shannon diversity values between individuals with normal colons and carcinomas. Blue points represent individuals with normal colons, yellow points represent individuals with adenomas (A), and red points represent individuals with carcinomas (B and C). The black lines represent the median value for each group.
FIG 2
Comparison of odds ratios calculated using alpha diversity community metrics associated with the presence of adenomas (A) or carcinomas (B) relative to those in individuals with normal colons using data collected from stool samples.
FIG 3
AUC values when classifying individuals as having normal colons or carcinomas using taxa with significant ORs when using stool samples (A) and unmatched tissue samples (B). We did not identify any taxa as having a significant OR to differentiate individuals with normal colons and adenomas or using matched tissue samples. The large black circles represent the median AUC of all studies, and the smaller circles represent the individual AUC for a particular study. The dashed line denotes an AUC of 0.5.
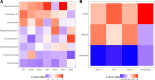
FIG 4
Relative importance of taxa with significant ORs in Random Forest models for differentiating between individuals with normal colons and carcinomas using stool samples (A) or unmatched tissue samples (B). The colors indicate the Z-transformed (i.e., mean of 0.0 and standard deviation of 1.0) mean decrease in accuracy values calculated from the model for each study. The taxa are ranked by their mean Z-score-transformed mean decrease in accuracy.
FIG 5
Comparison of Random Forest modeling approaches to classify individuals as having normal colons or adenomas (A) or carcinomas (B) when training the models using the taxa with significant ORs, all taxa in a community, or all OTUs in a community when using stool samples. No taxon had a significant OR associated with the presence of adenomas using stool samples. The black line represents the median AUC for the respective group. The dashed gray line indicates an AUC of 0.5.
FIG 6
Testing of Random Forest models to classify individuals as having normal colons or adenomas (A) or carcinomas (B) when using sequence data obtained from stool samples. Models were trained on data from each study (Fig. 5) and tested on the other studies. The black lines represent the median AUC of all test AUCs for a specific study. The dashed gray line represents the AUC at 0.5.
Similar articles
- Normalization of the microbiota in patients after treatment for colonic lesions.
Sze MA, Baxter NT, Ruffin MT 4th, Rogers MAM, Schloss PD. Sze MA, et al. Microbiome. 2017 Nov 16;5(1):150. doi: 10.1186/s40168-017-0366-3. Microbiome. 2017. PMID: 29145893 Free PMC article. - Fecal Short-Chain Fatty Acids Are Not Predictive of Colonic Tumor Status and Cannot Be Predicted Based on Bacterial Community Structure.
Sze MA, Topçuoğlu BD, Lesniak NA, Ruffin MT 4th, Schloss PD. Sze MA, et al. mBio. 2019 Jul 2;10(4):e01454-19. doi: 10.1128/mBio.01454-19. mBio. 2019. PMID: 31266879 Free PMC article. - Re-purposing 16S rRNA gene sequence data from within case paired tumor biopsy and tumor-adjacent biopsy or fecal samples to identify microbial markers for colorectal cancer.
Shah MS, DeSantis T, Yamal JM, Weir T, Ryan EP, Cope JL, Hollister EB. Shah MS, et al. PLoS One. 2018 Nov 9;13(11):e0207002. doi: 10.1371/journal.pone.0207002. eCollection 2018. PLoS One. 2018. PMID: 30412600 Free PMC article. - Robust prediction of colorectal cancer via gut microbiome 16S rRNA sequencing data.
Porreca A, Ibrahimi E, Maturo F, Marcos Zambrano LJ, Meto M, Lopes MB. Porreca A, et al. J Med Microbiol. 2024 Oct;73(10). doi: 10.1099/jmm.0.001903. J Med Microbiol. 2024. PMID: 39377779 - Valid and accepted novel bacterial taxa isolated from non-domestic animals and taxonomic revisions published in 2023.
Munson E, Burbick CR, Lawhon SD, Krueger T, Ruiz-Reyes E. Munson E, et al. J Clin Microbiol. 2024 Oct 16;62(10):e0104224. doi: 10.1128/jcm.01042-24. Epub 2024 Oct 1. J Clin Microbiol. 2024. PMID: 39352133 Review.
Cited by
- Leveraging Fecal Bacterial Survey Data to Predict Colorectal Tumors.
Zhang B, Xu S, Xu W, Chen Q, Chen Z, Yan C, Fan Y, Zhang H, Liu Q, Yang J, Yang J, Xiao C, Xu H, Ren J. Zhang B, et al. Front Genet. 2019 May 28;10:447. doi: 10.3389/fgene.2019.00447. eCollection 2019. Front Genet. 2019. PMID: 31191599 Free PMC article. - Statistical and Machine Learning Techniques in Human Microbiome Studies: Contemporary Challenges and Solutions.
Moreno-Indias I, Lahti L, Nedyalkova M, Elbere I, Roshchupkin G, Adilovic M, Aydemir O, Bakir-Gungor B, Santa Pau EC, D'Elia D, Desai MS, Falquet L, Gundogdu A, Hron K, Klammsteiner T, Lopes MB, Marcos-Zambrano LJ, Marques C, Mason M, May P, Pašić L, Pio G, Pongor S, Promponas VJ, Przymus P, Saez-Rodriguez J, Sampri A, Shigdel R, Stres B, Suharoschi R, Truu J, Truică CO, Vilne B, Vlachakis D, Yilmaz E, Zeller G, Zomer AL, Gómez-Cabrero D, Claesson MJ. Moreno-Indias I, et al. Front Microbiol. 2021 Feb 22;12:635781. doi: 10.3389/fmicb.2021.635781. eCollection 2021. Front Microbiol. 2021. PMID: 33692771 Free PMC article. - The human oral - nasopharynx microbiome as a risk screening tool for nasopharyngeal carcinoma.
Hao Y, Zeng Z, Peng X, Ai P, Han Q, Ren B, Li M, Wang H, Zhou X, Zhou X, Ma Y, Cheng L. Hao Y, et al. Front Cell Infect Microbiol. 2022 Nov 30;12:1013920. doi: 10.3389/fcimb.2022.1013920. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36530430 Free PMC article. - Recent advancements in the exploitation of the gut microbiome in the diagnosis and treatment of colorectal cancer.
Stott KJ, Phillips B, Parry L, May S. Stott KJ, et al. Biosci Rep. 2021 Jul 30;41(7):BSR20204113. doi: 10.1042/BSR20204113. Biosci Rep. 2021. PMID: 34236075 Free PMC article. Review. - Library Preparation and Sequencing Platform Introduce Bias in Metagenomic-Based Characterizations of Microbiomes.
Poulsen CS, Ekstrøm CT, Aarestrup FM, Pamp SJ. Poulsen CS, et al. Microbiol Spectr. 2022 Apr 27;10(2):e0009022. doi: 10.1128/spectrum.00090-22. Epub 2022 Mar 15. Microbiol Spectr. 2022. PMID: 35289669 Free PMC article.
References
- Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, Casero RA. 2011. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A 108:15354–15359. doi:10.1073/pnas.1010203108. - DOI - PMC - PubMed
- Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. 2016. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20:215–225. doi:10.1016/j.chom.2016.07.006. - DOI - PMC - PubMed
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123. doi:10.1126/science.1224820. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials