Current approaches for the fitting and refinement of atomic models into cryo-EM maps using CCP-EM - PubMed (original) (raw)
Review
Current approaches for the fitting and refinement of atomic models into cryo-EM maps using CCP-EM
Robert A Nicholls et al. Acta Crystallogr D Struct Biol. 2018.
Abstract
Recent advances in instrumentation and software have resulted in cryo-EM rapidly becoming the method of choice for structural biologists, especially for those studying the three-dimensional structures of very large macromolecular complexes. In this contribution, the tools available for macromolecular structure refinement into cryo-EM reconstructions that are available via CCP-EM are reviewed, specifically focusing on REFMAC5 and related tools. Whilst originally designed with a view to refinement against X-ray diffraction data, some of these tools have been able to be repurposed for cryo-EM owing to the same principles being applicable to refinement against cryo-EM maps. Since both techniques are used to elucidate macromolecular structures, tools encapsulating prior knowledge about macromolecules can easily be transferred. However, there are some significant qualitative differences that must be acknowledged and accounted for; relevant differences between these techniques are highlighted. The importance of phases is considered and the potential utility of replacing inaccurate amplitudes with their expectations is justified. More pragmatically, an upper bound on the correlation between observed and calculated Fourier coefficients, expressed in terms of the Fourier shell correlation between half-maps, is demonstrated. The importance of selecting appropriate levels of map blurring/sharpening is emphasized, which may be facilitated by considering the behaviour of the average map amplitude at different resolutions, as well as the utility of simultaneously viewing multiple blurred/sharpened maps. Features that are important for the purposes of computational efficiency are discussed, notably the Divide and Conquer pipeline for the parallel refinement of large macromolecular complexes. Techniques that have recently been developed or improved in Coot to facilitate and expedite the building, fitting and refinement of atomic models into cryo-EM maps are summarized. Finally, a tool for symmetry identification from a given map or coordinate set, ProSHADE, which can identify the point group of a map and thus may be used during deposition as well as during molecular visualization, is introduced.
Keywords: Divide and Conquer; ProSHADE; REFMAC5; cryo-EM; map blurring; map sharpening; model refinement; symmetry detection.
open access.
Figures
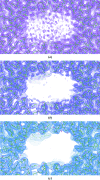
Figure 1
The effect of map blurring on cryo-EM reconstructions. Electrostatic potential maps and atomic models corresponding to the structure of β-galactosidase (PDB entry
5a1a
, EMDB entry EMD-2084; Bartesaghi et al., 2015 ▸). (a) The original map is very noisy, suggesting that it is over-sharpened. (b) Blurring the map using a B value of 40 Å reduces the noise, appearing to make it largely disappear, although the features (signal) in the map remain clear. This produces a clearer map that it will be easier to refine the atomic model against. (c) Applying further blurring (B value of 100 Å) does not help: it causes more of the noise to disappear, but also causes structural details to be lost (for example regarding side chains).
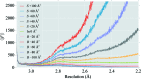
Figure 2
Plot of average structure-factor amplitude against resolution to aid the selection of the optimal blurring/sharpening B value, as shown in the CCP-EM GUI (Burnley et al., 2017 ▸). The data correspond to a cryo-EM reconstruction of β-galactosidase (PDB entry
5a1a
, EMDB entry EMD-2084; Bartesaghi et al., 2015 ▸). Different levels of map blurring and sharpening are shown, using an array of B values from −100 Å (blurring) to +100 Å (sharpening) in increments of 20 Å. For the optimally blurred/sharpened map, the average amplitude should decay to zero at the ‘high-resolution limit’ (above which the data contain no information about the true structure). For high levels of map sharpening, the average amplitude persistently increases with resolution: this indicates over-sharpening. For high levels of map blurring, the average amplitude approaches zero rapidly prior to reaching the high-resolution limit, resulting in a loss of high-resolution information. In this case, the optimal average level of blurring over the map is around 40 Å (subjectively determined by manual inspection of the plot), in agreement with the maps shown in Figs. 1 ▸ and 3 ▸.
Figure 3
The utility of simultaneously viewing multiple maps using various levels of map blurring/sharpening. Electrostatic potential maps and atomic models corresponding to β-galactosidase (PDB entry
5a1a
, EMDB entry EMD-2084; Bartesaghi et al., 2015 ▸) using different levels of map blurring. (a) In the original/default map, noise is visible in the solvent regions and there is little backbone density visible in the exposed region around residue 732 in chain A. (b) Blurring the map using a B value of 40 Å reduces noise and reveals features in the map. This suggests incorrect modelling of the backbone in this region and indicates where the model backbone should be positioned. (c) Simultaneously viewing an array of maps with different levels of sharpening and blurring further increases map interpretability. Here, maps corresponding to blurring B values of 0, 20, 40, 60, 80 and 100 Å are displayed. In this case, it is evident how the backbone should be traced (from the more blurred maps), and at the same time it is possible to interpret finer structural details such as the orientations of side chains (from the sharper maps).
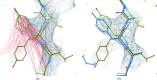
Figure 4
External restraint visualization and usage during parallelized full-chain real-space atomic model refinement in Coot. In order to mimic a ‘typical’ stage of the model-building and refinement process in which the model is complete yet the conformational geometry is suboptimal, the atomic model corresponding to the RAD51 filament (PDB entry
5jzc
, EMDB entry EMD-8183; Short et al., 2016 ▸) was subjected to simple re-refinement, causing overfitting into the map. External restraints were generated by ProSMART using a homologous structural model derived using MX (PDB entry
1n0w
; Pellegrini et al., 2002 ▸) as a reference. In Coot, the colouring of interatomic vectors representing restraints indicates local structural conservation; consistent interatomic distances are coloured grey, whilst those that are substantially longer/shorter in the target than in the reference model are coloured red/blue. (a) The re-refined model of
5jzc
is shown, along with the interatomic distance restraints that were generated using
1n0w
. From observing the restraint colouring it is evident that there are substantial local conformational differences between the two structural models despite the sequence homology. (b) Performing full-chain real-space refinement of the model in Coot results in the local conformation of the model becoming much closer to that of the high-resolution homologue, as indicated by the increased quantity of interatomic vectors coloured grey. Regions containing restraints coloured red/blue indicate persisting differences between the target and reference models. Such regions are indicative of either true differences between the target and reference structures (as is the case in Fig. 5 ▸), suboptimal restraint weighting parameters or errors in the model that require manual resolution. Regardless, the presence of any such regions would warrant closer inspection.
Figure 5
Robustness to outliers when performing atomic model refinement using external restraints. The re-refined atomic model corresponding to the RAD51 filament (PDB entry
5jzc
, EMDB entry EMD-8183; Short et al., 2016 ▸), as shown in Fig. 4 ▸(a), is displayed coloured green. External restraints were generated using ProSMART using the homologous structural model derived using MX (PDB entry
1n0w
; Pellegrini et al., 2002 ▸) as a reference. The homologous model
1n0w
is shown coloured brown superposed onto
5jzc
. (a) The red colouring of the restraints reflects differences in the rotameric state of the aligned functional tyrosine between the target and reference models (Tyr293 in
5jzc
, Tyr301 in
1n0w
). Map density is clearly visible; this side chain is oriented acceptably in the original model, so it would be undesirable for the external restraints to pull this side chain into the conformation observed in the homologous model. (b) The model after real-space re-refinement with external restraints in Coot (as shown in Fig. 4 ▸b) is coloured blue. Owing to the use of robust estimation, which restricts the influence of outliers, the tyrosine side chain retains its original conformation rather than being pulled out of the map and into the rotameric state observed in the homologous model. However, the geometry of the backbone in this region is improved, adopting a similar conformation to the homologous model as appropriate. Following such injection of prior structural information, the typical protocol would then be to perform full-model refinement using _REFMAC_5 with jelly-body restraints, which would allow the model to relax into the density whilst ensuring refinement stability.
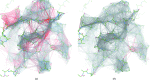
Figure 6
The Divide and Conquer procedure illustrated using a model of a rotavirus particle (PDB entry
4v7q
, EMDB entry EMD-5199; Settembre et al., 2011 ▸) visualized using PyMOL (Schrödinger). (a) The input macromolecule is split into individual chains. (b) For each chain, the adjacent contacting chains are identified and included for the refinement of the target chain. Each such substructure is masked in order to identify an appropriate box size for use in refinement. (c) After parallel refinement of each individual substructure, all of the target chains are extracted and recombined into the macromolecular complex.
Figure 7
Symmetry detection using ProSHADE. The map corresponding to a bacteriophage T4 portal protein (PDB entry
3ja7
, EMDB entry EMD-6324; Sun et al., 2015 ▸) visualized using UCSF Chimera (Pettersen et al., 2004 ▸). ProSHADE reports the correct _C_12 symmetry, with the symmetry axis along the vector (0.02, 0.02, 1.00) with an average cross-correlation between the original structure and the structures rotated along this symmetry axis of 0.443. All symmetry elements present for this instance of the _C_12 symmetry group are reported, along with the list of alternative symmetries that are also present in the structure (_C_6, _C_4, _C_3 and _C_2) and the corresponding rotation-function peak heights.
Similar articles
- Refinement of Atomic Structures Against cryo-EM Maps.
Murshudov GN. Murshudov GN. Methods Enzymol. 2016;579:277-305. doi: 10.1016/bs.mie.2016.05.033. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27572731 Review. - Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions.
Brown A, Long F, Nicholls RA, Toots J, Emsley P, Murshudov G. Brown A, et al. Acta Crystallogr D Biol Crystallogr. 2015 Jan 1;71(Pt 1):136-53. doi: 10.1107/S1399004714021683. Epub 2015 Jan 1. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25615868 Free PMC article. - Cryo-EM single-particle structure refinement and map calculation using Servalcat.
Yamashita K, Palmer CM, Burnley T, Murshudov GN. Yamashita K, et al. Acta Crystallogr D Struct Biol. 2021 Oct 1;77(Pt 10):1282-1291. doi: 10.1107/S2059798321009475. Epub 2021 Sep 29. Acta Crystallogr D Struct Biol. 2021. PMID: 34605431 Free PMC article. - Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data.
Casañal A, Lohkamp B, Emsley P. Casañal A, et al. Protein Sci. 2020 Apr;29(4):1069-1078. doi: 10.1002/pro.3791. Epub 2020 Mar 2. Protein Sci. 2020. PMID: 31730249 Free PMC article. - Overview and applications of map and model validation tools in the CCP-EM software suite.
Joseph AP, Malhotra S, Burnley T, Winn MD. Joseph AP, et al. Faraday Discuss. 2022 Nov 8;240(0):196-209. doi: 10.1039/d2fd00103a. Faraday Discuss. 2022. PMID: 35916020 Free PMC article. Review.
Cited by
- Community recommendations on cryoEM data archiving and validation: Outcomes of a wwPDB/EMDB workshop on cryoEM data management, deposition and validation.
Kleywegt GJ, Adams PD, Butcher SJ, Lawson CL, Rohou A, Rosenthal PB, Subramaniam S, Topf M, Abbott S, Baldwin PR, Berrisford JM, Bricogne G, Choudhary P, Croll TI, Danev R, Ganesan SJ, Grant T, Gutmanas A, Henderson R, Heymann JB, Huiskonen JT, Istrate A, Kato T, Lander GC, Lok SM, Ludtke SJ, Murshudov GN, Pye R, Pintilie GD, Richardson JS, Sachse C, Salih O, Scheres SHW, Schroeder GF, Sorzano COS, Stagg SM, Wang Z, Warshamanage R, Westbrook JD, Winn MD, Young JY, Burley SK, Hoch JC, Kurisu G, Morris K, Patwardhan A, Velankar S. Kleywegt GJ, et al. ArXiv [Preprint]. 2024 Feb 2:arXiv:2311.17640v3. ArXiv. 2024. PMID: 38076521 Free PMC article. Updated. Preprint. - Dual species sphingosine-1-phosphate lyase inhibitors to combine antifungal and anti-inflammatory activities in cystic fibrosis: a feasibility study.
Cellini B, Pampalone G, Camaioni E, Pariano M, Catalano F, Zelante T, Dindo M, Macchioni L, Di Veroli A, Galarini R, Paoletti F, Davidescu M, Stincardini C, Vascelli G, Bellet MM, Saba J, Giovagnoli S, Giardina G, Romani L, Costantini C. Cellini B, et al. Sci Rep. 2023 Dec 20;13(1):22692. doi: 10.1038/s41598-023-50121-4. Sci Rep. 2023. PMID: 38123809 Free PMC article. - Haruspex: A Neural Network for the Automatic Identification of Oligonucleotides and Protein Secondary Structure in Cryo-Electron Microscopy Maps.
Mostosi P, Schindelin H, Kollmannsberger P, Thorn A. Mostosi P, et al. Angew Chem Int Ed Engl. 2020 Aug 24;59(35):14788-14795. doi: 10.1002/anie.202000421. Epub 2020 May 11. Angew Chem Int Ed Engl. 2020. PMID: 32187813 Free PMC article. - Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
Seyfizadeh N, Kalbermatter D, Imhof T, Ries M, Müller C, Jenner L, Blumenschein E, Yendrzheyevskiy A, Grün F, Moog K, Eckert D, Engel R, Diebolder P, Chami M, Krauss J, Schaller T, Arndt M. Seyfizadeh N, et al. J Biomed Sci. 2024 May 28;31(1):56. doi: 10.1186/s12929-024-01045-2. J Biomed Sci. 2024. PMID: 38807208 Free PMC article. - Theoretical 3D electron diffraction electrostatic potential maps of proteins modeled with a multipolar pseudoatom data bank.
Kulik M, Chodkiewicz ML, Dominiak PM. Kulik M, et al. Acta Crystallogr D Struct Biol. 2022 Aug 1;78(Pt 8):1010-1020. doi: 10.1107/S2059798322005836. Epub 2022 Jul 14. Acta Crystallogr D Struct Biol. 2022. PMID: 35916225 Free PMC article.
References
- Afonine, P. V., Headd, J. J., Terwilliger, T. C. & Adams, P. D. (2013). Comput. Crystallogr. Newsl. 4, 43–44. https://www.phenix-online.org/newsletter/CCN_2013_07.pdf.
Publication types
MeSH terms
Substances
Grants and funding
- MC_UP_A025_1012/MRC_/Medical Research Council/United Kingdom
- MC_US_A025_1012/MRC_/Medical Research Council/United Kingdom
- BB/L007010/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- PR140014/Science and Technology Facilities Council
LinkOut - more resources
Full Text Sources
Other Literature Sources