Cancer Drug Response Profile scan (CDRscan): A Deep Learning Model That Predicts Drug Effectiveness from Cancer Genomic Signature - PubMed (original) (raw)
Cancer Drug Response Profile scan (CDRscan): A Deep Learning Model That Predicts Drug Effectiveness from Cancer Genomic Signature
Yoosup Chang et al. Sci Rep. 2018.
Abstract
In the era of precision medicine, cancer therapy can be tailored to an individual patient based on the genomic profile of a tumour. Despite the ever-increasing abundance of cancer genomic data, linking mutation profiles to drug efficacy remains a challenge. Herein, we report Cancer Drug Response profile scan (CDRscan) a novel deep learning model that predicts anticancer drug responsiveness based on a large-scale drug screening assay data encompassing genomic profiles of 787 human cancer cell lines and structural profiles of 244 drugs. CDRscan employs a two-step convolution architecture, where the genomic mutational fingerprints of cell lines and the molecular fingerprints of drugs are processed individually, then merged by 'virtual docking', an in silico modelling of drug treatment. Analysis of the goodness-of-fit between observed and predicted drug response revealed a high prediction accuracy of CDRscan (R2 > 0.84; AUROC > 0.98). We applied CDRscan to 1,487 approved drugs and identified 14 oncology and 23 non-oncology drugs having new potential cancer indications. This, to our knowledge, is the first-time application of a deep learning model in predicting the feasibility of drug repurposing. By further clinical validation, CDRscan is expected to allow selection of the most effective anticancer drugs for the genomic profile of the individual patient.
Conflict of interest statement
The authors declare no competing interests.
Figures
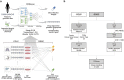
Figure 1
Overview of Cancer Drug Response profile scan (CDRscan). (a) Two main applications of CDRscan and dataset structure. For any given genomic fingerprint (i.e., a list of somatic mutations) of a tumour, CDRscan predicts which of 244 Genomics in Drug Sensitivity in Cancer (GDSC) anticancer drugs would be effective. The input of CDRscan can be molecular information of a particular small molecule for which CDRscan reports the predicted sensitivity of 787 cancer cell lines. The datasets used to train CDRscan were extracted from COSMIC cell line project (CCLP) and GDSC databases which represent 787 cancer cell lines across 25 cancer types defined by TCGA, 28,328 mutation positions in 567 cancer associated genes, and assay results from treatment of 244 anticancer drugs. (b) Data filtering procedure and final datasets. CCLP and GDSC databases contain genomic characterisation of 1,001 cancer cell lines and IC50 values measured from treatment of 1,001 cell lines with 265 anticancer drugs. The datasets were refined to include only the 567 Cosmic Cancer Gene Census genes and the cancer types that have at least 10 cell lines. Drugs without PubChem Compound Identifier or having molecular weight greater than 1000 g/mol were excluded. Totals of 28,328 and 3,072 features were extracted from cell line genomic signatures and drugs, respectively, constituting binary encoding of 31,400 features in total. The graphical image used in Fig. 1a is an original creation by Ye-Bin Jung and is reprinted under a CC BY license with permission from Ye-Bin Jung. All rights reserved.
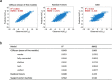
Figure 2
Assessment of prediction accuracy of CDRscan. (a) Scatter plots showing correlation between the observed and predicted IC50 values for CDRscan and two other machine learning models to benchmark the prediction accuracy. The test datasets, which correspond to 5% of the total cell line-drug pairs, were used to assess the coefficient of determination (R2). (b) Table summarizing the R2 values and root mean squared errors (RMSE) of CDRscan (mean value of the five models and values for individual models), random forest, and support vector machine.
Figure 3
Cell line- and drug-centric correlation analyses. (a) Prediction accuracy assessment for each cell line. Scatter plots show the correlation between observed and CDRscan-predicted ln(IC50) values for the cell lines that showed the strongest (BFTC-909, left) and the lowest agreement (COR-L32, right). The COSMIC IDs of the two cell lines and the corresponding cancer types are indicated above the scatter plots, and the R2 values, Pearson correlation coefficient (r), p values, and the number of instances (n) are shown in the upper left corner of each plot. Histograms on the right show the overall distribution of prediction accuracy assessed for individual cell lines using indicated metrics. (b) Scatter plots showing the strongest and weakest agreement between observed and CDRscan-predicted ln(IC50) in drug-centric correlation analysis. The drug name and its PubCHEM ID are indicated in each plot. The R2 values, Pearson correlation coefficient (r), p values, and the instance counts (n) are also indicated. Histograms on the right show the overall distribution of prediction accuracy (R2) assessed for individual drugs using indicated metrics.
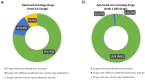
Figure 4
Feasibility of drug repurposing using CDRscan (a) Approved anticancer drugs with potential repurposing opportunity. CDRscan predicted that 23 out of 102 approved anticancer drugs have activity against at least one new cancer type in addition to the originally approved indications. Nine of these showed predictive sensitivity of more than 90% cancer types, indicating nonspecific antiproliferative/cytotoxicitc effects. (b) Approved non-oncology drugs with potential repurposing opportunity. Of the 1,385 non-oncology drugs, 27 showed potential anticancer activity. Four of these 27 drugs were predicted to have activity against over 90% of cancer types.
Similar articles
- DeepDRK: a deep learning framework for drug repurposing through kernel-based multi-omics integration.
Wang Y, Yang Y, Chen S, Wang J. Wang Y, et al. Brief Bioinform. 2021 Sep 2;22(5):bbab048. doi: 10.1093/bib/bbab048. Brief Bioinform. 2021. PMID: 33822890 - RefDNN: a reference drug based neural network for more accurate prediction of anticancer drug resistance.
Choi J, Park S, Ahn J. Choi J, et al. Sci Rep. 2020 Feb 5;10(1):1861. doi: 10.1038/s41598-020-58821-x. Sci Rep. 2020. PMID: 32024872 Free PMC article. - Improved anticancer drug response prediction in cell lines using matrix factorization with similarity regularization.
Wang L, Li X, Zhang L, Gao Q. Wang L, et al. BMC Cancer. 2017 Aug 2;17(1):513. doi: 10.1186/s12885-017-3500-5. BMC Cancer. 2017. PMID: 28768489 Free PMC article. - Computer-aided drug repurposing for cancer therapy: Approaches and opportunities to challenge anticancer targets.
Mottini C, Napolitano F, Li Z, Gao X, Cardone L. Mottini C, et al. Semin Cancer Biol. 2021 Jan;68:59-74. doi: 10.1016/j.semcancer.2019.09.023. Epub 2019 Sep 25. Semin Cancer Biol. 2021. PMID: 31562957 Review. - Machine and deep learning approaches for cancer drug repurposing.
Issa NT, Stathias V, Schürer S, Dakshanamurthy S. Issa NT, et al. Semin Cancer Biol. 2021 Jan;68:132-142. doi: 10.1016/j.semcancer.2019.12.011. Epub 2020 Jan 3. Semin Cancer Biol. 2021. PMID: 31904426 Free PMC article. Review.
Cited by
- Understanding Drug Repurposing From the Perspective of Biomedical Entities and Their Evolution: Bibliographic Research Using Aspirin.
Li X, Rousseau JF, Ding Y, Song M, Lu W. Li X, et al. JMIR Med Inform. 2020 Jun 16;8(6):e16739. doi: 10.2196/16739. JMIR Med Inform. 2020. PMID: 32543442 Free PMC article. - Machine learning and feature selection for drug response prediction in precision oncology applications.
Ali M, Aittokallio T. Ali M, et al. Biophys Rev. 2019 Feb;11(1):31-39. doi: 10.1007/s12551-018-0446-z. Epub 2018 Aug 10. Biophys Rev. 2019. PMID: 30097794 Free PMC article. Review. - Functional regulations between genetic alteration-driven genes and drug target genes acting as prognostic biomarkers in breast cancer.
Wang L, Yu L, Shi J, Li F, Zhang C, Xu H, Yin X, Wang L, Lin S, Litvinova A, Ping Y, Ning S, Zhao H. Wang L, et al. Sci Rep. 2022 Jun 23;12(1):10641. doi: 10.1038/s41598-022-13835-5. Sci Rep. 2022. PMID: 35739271 Free PMC article. - Deep reinforcement learning for personalized treatment recommendation.
Liu M, Shen X, Pan W. Liu M, et al. Stat Med. 2022 Sep 10;41(20):4034-4056. doi: 10.1002/sim.9491. Epub 2022 Jun 18. Stat Med. 2022. PMID: 35716038 Free PMC article. - Global development of artificial intelligence in cancer field: a bibliometric analysis range from 1983 to 2022.
Wang SH, Chen G, Zhong X, Lin T, Shen Y, Fan X, Cao L. Wang SH, et al. Front Oncol. 2023 Jul 14;13:1215729. doi: 10.3389/fonc.2023.1215729. eCollection 2023. Front Oncol. 2023. PMID: 37519796 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources