The gut microbiota metabolite indole alleviates liver inflammation in mice - PubMed (original) (raw)
. 2018 Jun 15;32(12):fj201800544.
doi: 10.1096/fj.201800544. Online ahead of print.
Audrey M Neyrinck 1, Marta Olivares 1, Julie Rodriguez 1, Audrey de Rocca Serra 2, Martin Roumain 3, Laure B Bindels 1, Patrice D Cani 1 4, Pieter Evenepoel 5, Giulio G Muccioli 3, Jean-Baptiste Demoulin 2, Nathalie M Delzenne 1
Affiliations
- PMID: 29906245
- PMCID: PMC6219839
- DOI: 10.1096/fj.201800544
The gut microbiota metabolite indole alleviates liver inflammation in mice
Martin Beaumont et al. FASEB J. 2018.
Abstract
The gut microbiota regulates key hepatic functions, notably through the production of bacterial metabolites that are transported via the portal circulation. We evaluated the effects of metabolites produced by the gut microbiota from aromatic amino acids (phenylacetate, benzoate, p-cresol, and indole) on liver inflammation induced by bacterial endotoxin. Precision-cut liver slices prepared from control mice, Kupffer cell (KC)-depleted mice, and obese mice ( ob/ ob) were treated with or without LPS and bacterial metabolites. We observed beneficial effects of indole that dose-dependently reduced the LPS-induced up-regulation of proinflammatory mediators at both mRNA and protein levels in precision-cut liver slices prepared from control or ob/ ob mice. KC depletion partly prevented the antiinflammatory effects of indole, notably through a reduction of nucleotide-binding domain and leucine-rich repeat containing (NLR) family pyrin domain-containing 3 (NLRP3) pathway activation. In vivo, the oral administration of indole before an LPS injection reduced the expression of key proteins of the NF-κB pathway and downstream proinflammatory gene up-regulation. Indole also prevented LPS-induced alterations of cholesterol metabolism through a transcriptional regulation associated with increased 4β-hydroxycholesterol hepatic levels. In summary, indole appears as a bacterial metabolite produced from tryptophan that is able to counteract the detrimental effects of LPS in the liver. Indole could be a new target to develop innovative strategies to decrease hepatic inflammation.-Beaumont, M., Neyrinck, A. M., Olivares, M., Rodriguez, J., de Rocca Serra, A., Roumain, M., Bindels, L. B., Cani, P. D., Evenepoel, P., Muccioli, G. G., Demoulin, J.-B., Delzenne, N. M. The gut microbiota metabolite indole alleviates liver inflammation in mice.
Keywords: Kupffer cells; LPS; PCLS; cholesterol metabolism; gut–liver axis.
Conflict of interest statement
The authors thank V. Allaeys and B. Es Saadi (Metabolism and Nutrition Research Group, Louvain Drug Research Institute) for their skillful technical assistance. N.M.D. is a recipient of an European Union (EU) grant (613979 MyNewGut Project) and Belgium National Scientific Research Fund (FRS-FNRS) (Belgium) grants. M.O. is a beneficiary of a “MOVE-IN Louvain” Incoming Postdoctoral Fellowship cofunded by the Marie Curie Actions of the European Commission. P.D.C., a senior research associate at the FRS-FNRS, is a recipient of a 2013 European Research Council Starting grant (336452-ENIGMO) and a 2015 Baillet Latour grant for medical research, and is supported by the FRS-FNRS via the Fund for Strategic Fundamental Research (FRFS)–Walloon Excellence in Life Sciences and Biotechnology (WELBIO) under Grant WELBIO-CGR-2017. A.D.R.S. is a recipient of a postdoctoral fellowship from FRS-FNRS. The authors declare no conflicts of interest.
Figures
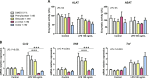
Figure 1
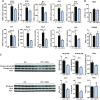
Effects of bacterial metabolites produced from AAA on PCLS. PCLS were treated with or without LPS (100 ng/ml) and bacterial metabolites (1 mM). A) ALAT and ASAT activities were measured in culture medium. B) mRNA levels were quantified in PCLS using real-time quantitative PCR. Data are presented as means ±
sem
, n ≥ 4 mice in each condition (experiment 1). Mixed-model ANOVA was used with LPS and metabolite treatments as fixed effects and mouse as random effect. For pairwise comparison, mean value of each metabolite-treated group was compared to mean of respective vehicle-treated group in water or LPS conditions. NS, not significant. ***P < 0.001 (adjusted by Tukey method).
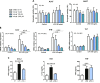
Figure 2
Dose effects of indole on PCLS. PCLS were treated with or without LPS (100 ng/ml) and indole (10–1000 µM). A) ALAT and ASAT were measured in culture medium. B) mRNA levels were quantified in PCLS using real-time quantitative PCR. C) Cytokines were quantified in culture medium using Luminex. For this experiment, intermediate concentration of indole was used (100 µM). For IL-1B, concentration in control and LPS + indole group were extrapolated beyond standard range, but fluorescent signal was always higher than signal measured in blank. Data are presented as means ±
sem
, n ≥ 5 mice in each condition (experiment 1). Mixed-model ANOVA was used with LPS and indole (I) treatments as fixed effects and mouse as random effect. For pairwise comparison, mean value of each indole-treated group was compared to mean of respective vehicle group in water- or LPS-treated conditions. NS, not significant. ***P < 0.001, **P < 0.01 *P < 0.05 (adjusted by Tukey method).
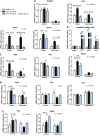
Figure 3
Effects of indole on PCLS prepared from mice without KC. PCLS were prepared after injection of CL or NaCl (control) before treatment with or without LPS (100 ng/ml) and indole (100 µM). A, C_–_E) mRNA levels were quantified in PCLS using real-time quantitative PCR, n ≥ 8 mice in each condition (experiment 2). Mixed-model ANOVA was used with CL, LPS, and indole treatments as fixed effects and mouse as random effect. Mean values were compared pairwise in control or CL conditions. B) mRNA levels were quantified by real-time quantitative PCR in isolated KC treated with or without LPS (100 ng/ml) and indole (100 µM), n = 4 mice in each condition (experiment 1). Mixed-model ANOVA was used with LPS and indole treatments as fixed effects and mouse as random effect. Mean values were compared pairwise. Data are presented as means ±
sem
. NS, not significant. ***P < 0.001, **P < 0.01, *P < 0.05 (adjusted by Tukey method).
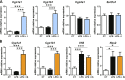
Figure 4
Interactions between indole and xenobiotic metabolism in PCLS. PCLS were treated with or without LPS (100 ng/ml) and indole or I3S (100 µM). mRNA levels were quantified by real-time quantitative PCR in PCLS treated with LPS and indole (A) or I3S (B) (experiment 1). Data are presented as means ±
sem
, n ≥ 4 mice in each condition. CT, control; I, indole; IS, I3S. Mixed-model ANOVA was used with LPS and metabolite treatments as fixed effects and mouse as random effect. Mean values were compared pairwise. ***P < 0.001, *P < 0.05 (adjusted by Tukey method).
Figure 5
Effects of indole on PCLS prepared from genetically obese (ob/ob) mice. PCLS were treated with or without indole (100 µM). mRNA levels were quantified in PCLS using real-time quantitative PCR. Data are presented as means ±
sem
, n ≥ 4 mice in each group (experiment 3). Mixed-model ANOVA was used with genotype and indole treatment as fixed effects and mouse as random effect. Mean values were compared pairwise in ob/+ or ob/ob groups. ***P < 0.001, **P < 0.01 (adjusted by Tukey method).
Figure 6
Effects of indole on liver inflammation in vivo. Mice orally received water or indole (3 µmol/20 g of body weight) before intraperitoneal injection of vehicle or LPS (10 mg/kg of body weight). A) mRNA levels were quantified in liver using real-time quantitative PCR. Results are expressed relative to mRNA level measured in control group (set at 1). B) ROS and TBARS were quantified in liver. C) Left: Western blot of β-actin, and total and phosphorylated forms of NF-κB p65 and IκBα in liver lysate. right: band intensities were quantified relative to band intensity of β-actin, used as loading control. Data are presented as means ±
sem
, n = 6 mice in LPS and LPS + indole (LPS + I) groups, n = 4 in control group (CT) (experiment 4). ANOVA was used to test treatment effect, and mean values were compared pairwise. ***P < 0.001, **P < 0.01, *P < 0.05 (adjusted by Tukey method).
Figure 7
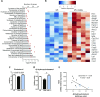
Effects of indole on liver sterols metabolism in vivo. Mice orally received water or indole (3 µmol per 20 g of body weight) before intraperitoneal injection of vehicle or LPS (10 mg/kg of body weight). Liver transcriptome was compared between LPS and LPS + indole (LPS + I) groups using microarrays (n = 5 mice in each group) (experiment 4). A) Significantly enriched functions implicated in biologic category “Lipid metabolism” identified by Ingenuity Pathway Analysis software. Bars show number of DEG implicated in each function. Red dots represent −log2 (P value) of enrichment. B) Heat map representing mRNA levels (log2) of selected DEG participating to functions enriched in biologic category, “Lipid metabolism.” C, D) Cholesterol (C) and 4β-hydroxycholesterol (D) were quantified in liver. Data are presented as means ±
sem
, n = 6 mice in LPS and LPS + I groups, n = 4 in control group. ANOVA was used to test treatment effect, and mean values were compared pairwise. *P < 0.05 (adjusted by Tukey method). E) Spearman correlation between 4β-hydroxycholesterol hepatic concentration and Nos2 mRNA levels in liver of mice treated with LPS (black dots, n = 6) or LPS + I (blue dots, n = 6).
Similar articles
- Hepatoprotective Effects of Indole, a Gut Microbial Metabolite, in Leptin-Deficient Obese Mice.
Knudsen C, Neyrinck AM, Leyrolle Q, Baldin P, Leclercq S, Rodriguez J, Beaumont M, Cani PD, Bindels LB, Lanthier N, Delzenne NM. Knudsen C, et al. J Nutr. 2021 Jun 1;151(6):1507-1516. doi: 10.1093/jn/nxab032. J Nutr. 2021. PMID: 33693866 Free PMC article. - Host microbiota dictates the proinflammatory impact of LPS in the murine liver.
Suriguga S, Luangmonkong T, Mutsaers HAM, Groothuis GMM, Olinga P. Suriguga S, et al. Toxicol In Vitro. 2020 Sep;67:104920. doi: 10.1016/j.tiv.2020.104920. Epub 2020 Jun 24. Toxicol In Vitro. 2020. PMID: 32590029 - A Novel Synbiotic Alleviates Autoimmune Hepatitis by Modulating the Gut Microbiota-Liver Axis and Inhibiting the Hepatic TLR4/NF-κB/NLRP3 Signaling Pathway.
Kang Y, Kuang X, Yan H, Ren P, Yang X, Liu H, Liu Q, Yang H, Kang X, Shen X, Tong M, Li L, Wang X, Guo L, Ma J, Zhang F, Fan W. Kang Y, et al. mSystems. 2023 Apr 27;8(2):e0112722. doi: 10.1128/msystems.01127-22. Epub 2023 Feb 16. mSystems. 2023. PMID: 36794950 Free PMC article. - Cooperation of liver cells in health and disease.
Kmieć Z. Kmieć Z. Adv Anat Embryol Cell Biol. 2001;161:III-XIII, 1-151. doi: 10.1007/978-3-642-56553-3. Adv Anat Embryol Cell Biol. 2001. PMID: 11729749 Review. - Activation of aryl hydrocarbon receptor (AhR) in Alzheimer's disease: role of tryptophan metabolites generated by gut host-microbiota.
Salminen A. Salminen A. J Mol Med (Berl). 2023 Mar;101(3):201-222. doi: 10.1007/s00109-023-02289-5. Epub 2023 Feb 9. J Mol Med (Berl). 2023. PMID: 36757399 Free PMC article. Review.
Cited by
- The gut microbiome and dietary fibres: implications in obesity, cardiometabolic diseases and cancer.
Delzenne NM, Bindels LB, Neyrinck AM, Walter J. Delzenne NM, et al. Nat Rev Microbiol. 2024 Oct 10. doi: 10.1038/s41579-024-01108-z. Online ahead of print. Nat Rev Microbiol. 2024. PMID: 39390291 Review. - Targeting hepatic macrophages for non-alcoholic fatty liver disease therapy.
Tian Y, Ni Y, Zhang T, Cao Y, Zhou M, Zhao C. Tian Y, et al. Front Cell Dev Biol. 2024 Sep 5;12:1444198. doi: 10.3389/fcell.2024.1444198. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39300994 Free PMC article. Review. - Digestive dynamics: Unveiling interplay between the gut microbiota and the liver in macronutrient metabolism and hepatic metabolic health.
Kandalgaonkar MR, Kumar V, Vijay-Kumar M. Kandalgaonkar MR, et al. Physiol Rep. 2024 Jun;12(12):e16114. doi: 10.14814/phy2.16114. Physiol Rep. 2024. PMID: 38886098 Free PMC article. Review. - Nutrition, gastrointestinal microorganisms and metabolites in mastitis occurrence and control.
Wang Y, Zhao Y, Tang X, Nan X, Jiang L, Wang H, Liu J, Yang L, Yao J, Xiong B. Wang Y, et al. Anim Nutr. 2024 Mar 16;17:220-231. doi: 10.1016/j.aninu.2024.01.010. eCollection 2024 Jun. Anim Nutr. 2024. PMID: 38800734 Free PMC article. Review.
References
- Strnad P., Tacke F., Koch A., Trautwein C. (2017) Liver—guardian, modifier and target of sepsis. Nat. Rev. Gastroenterol. Hepatol. 14, 55–66 - PubMed
- Cani P. D., Bibiloni R., Knauf C., Waget A., Neyrinck A. M., Delzenne N. M., Burcelin R. (2008) Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes 57, 1470–1481 - PubMed
- Cani P. D., Possemiers S., Van de Wiele T., Guiot Y., Everard A., Rottier O., Geurts L., Naslain D., Neyrinck A., Lambert D. M., Muccioli G. G., Delzenne N. M. (2009) Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2–driven improvement of gut permeability. Gut 58, 1091–1103 - PMC - PubMed
- Heymann F., Tacke F. (2016) Immunology in the liver—from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 13, 88–110 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous