Loss of RXFP2 and INSL3 genes in Afrotheria shows that testicular descent is the ancestral condition in placental mammals - PubMed (original) (raw)
Loss of RXFP2 and INSL3 genes in Afrotheria shows that testicular descent is the ancestral condition in placental mammals
Virag Sharma et al. PLoS Biol. 2018.
Abstract
Descent of testes from a position near the kidneys into the lower abdomen or into the scrotum is an important developmental process that occurs in all placental mammals, with the exception of five afrotherian lineages. Since soft-tissue structures like testes are not preserved in the fossil record and since key parts of the placental mammal phylogeny remain controversial, it has been debated whether testicular descent is the ancestral or derived condition in placental mammals. To resolve this debate, we used genomic data of 71 mammalian species and analyzed the evolution of two key genes (relaxin/insulin-like family peptide receptor 2 [RXFP2] and insulin-like 3 [INSL3]) that induce the development of the gubernaculum, the ligament that is crucial for testicular descent. We show that both RXFP2 and INSL3 are lost or nonfunctional exclusively in four afrotherians (tenrec, cape elephant shrew, cape golden mole, and manatee) that completely lack testicular descent. The presence of remnants of once functional orthologs of both genes in these afrotherian species shows that these gene losses happened after the split from the placental mammal ancestor. These "molecular vestiges" provide strong evidence that testicular descent is the ancestral condition, irrespective of persisting phylogenetic discrepancies. Furthermore, the absence of shared gene-inactivating mutations and our estimates that the loss of RXFP2 happened at different time points strongly suggest that testicular descent was lost independently in Afrotheria. Our results provide a molecular mechanism that explains the loss of testicular descent in afrotherians and, more generally, highlight how molecular vestiges can provide insights into the evolution of soft-tissue characters.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Fig 1. Gene-inactivating mutations in RXFP2 in four afrotherian species.
(A) The exon-intron structure of the coding region of the RXFP2 gene is shown as boxes (exons, drawn to scale) and lines (introns, not drawn to scale). A vertical red line/arrowhead indicates a frameshifting deletion/insertion, with the number of deleted/inserted bases given above. Stop codon mutations are shown as a black vertical line. Splice site mutations are indicated by the mutated dinucleotide. A blue vertical line indicates a frame-preserving deletion. Red boxes are exons that are either deleted or accumulated numerous mutations that destroy any sequence similarity. All inactivating mutations were validated by unassembled genome sequencing reads stored in the SRA. Elephant, rock hyrax, and aardvark have an intact RXFP2 gene and are not shown. A filled star indicates mutations that we confirmed by PCR and Sanger sequencing in the lesser hedgehog tenrec; the exon 17 frameshift was also found in the greater hedgehog tenrec (S5A and S5B Fig). An open star indicates mutations that we confirmed by PCR and sequencing in the dugong, the sister species of the manatee (S5C and S5D Fig). (B-I) Examples of inactivating mutations and their validation by unassembled SRA reads. RXFP2, relaxin/insulin-like family peptide receptor 2; SRA, Sequence Read Archive.
Fig 2. Gene-inactivating mutations and critical amino acid mutations in INSL3.
(A) Functional domains of the INSL3 protein and the exon-intron structure of the INSL3 gene. Inactivating mutations are as in Fig 1, frame-preserving insertions/deletions are shown as blue lines/arrowheads. Exons but not introns are drawn to scale. Elephant, rock hyrax, and aardvark have an intact INSL3 and are not shown. (B) While the cape golden mole does not exhibit any gene-inactivating mutations (A), an INSL3 protein alignment of the A- and B-chain shows mutations (red background) at amino acids that are critical for structure and function of the mature hormone (gray background) [–48]. Disulfide bonds between Cys residues are indicated by blue lines. Residues that are affected by frameshifting deletions in the underlying tenrec or cape elephant shrew nucleotide sequence are indicated by asterisks. For these two species, we ignored these frameshifts and used the ancestral reading frame. Species in red font are testicond. Note that elephant and rock hyrax have no mutations at any of the critical sites. INSL3, insulin-like 3.
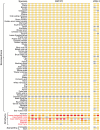
Fig 3. RXFP2 and INSL3 are only lost in several testicond afrotherian lineages.
The exon-intron structures of RXFP2 (18 coding exons) and INSL3 (2 coding exons) are shown. A yellow rectangle indicates an intact exon. Exons with inactivating mutations (stop codon or splice site mutations, frameshifts) are indicated in orange; completely deleted or highly diverged exons are indicated in red. Gray exons could not be examined because the respective genome assembly is incomplete at this locus (assembly gap). We could not examine the RXFP2 in the fragmented pangolin genome, because different parts of RXFP2 locus align to at least 5 different short scaffolds. Neither exons nor introns are drawn to scale. INSL3, insulin-like 3; RXFP2, relaxin/insulin-like family peptide receptor 2.
Fig 4. Evolution of RXFP2 and estimated time intervals during which RXFP2 came under neutral evolution.
Red boxes represent a lower and upper bound for an estimated time interval during which RXFP2 started to evolve neutrally (Materials and methods, S2 Table), which shows that the gene was lost independently at different time points. Gray boxes represent an estimated interval for species divergence times taken from TimeTree [57] (see also S4 Table). Species in blue font are testicond. The phylogenetic position of tenrecs, golden moles, elephant shrews is well supported by morphological and molecular characters [23, 24, 26]. Uncertain phylogenetic relationships are shown as a polytomy. Note that while the loss of RXFP2 in the elephant shrew is estimated to have happened at the base of the lineage, this loss is clearly independent from the much later loss in the tenrec and in the golden mole lineage. Evidence for the loss of RXFP2 in the dugong and the greater hedgehog tenrec was obtained by PCR and sequencing experiments (S5 Fig). RXFP2, relaxin/insulin-like family peptide receptor 2.
Similar articles
- Therian origin of INSL3/RXFP2-driven testicular descent in mammals.
Menzies BR, Tarulli GA, Frankenberg SR, Pask AJ. Menzies BR, et al. Front Cell Dev Biol. 2024 Feb 2;12:1353598. doi: 10.3389/fcell.2024.1353598. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38380341 Free PMC article. - An insight into insulin-like factor 3 regulate its receptor RXFP2 in mouse gubernaculum testis cells.
Duan S, Zhang X, Jiang X, Xie L, Sun Z, Ma S, Li J. Duan S, et al. Int J Clin Exp Pathol. 2015 Nov 1;8(11):14806-11. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26823808 Free PMC article. - Expression analyses of insulin-like peptide 3, RXFP2, LH receptor, and 3β-hydroxysteroid dehydrogenase in testes of normal and cryptorchid dogs.
Hannan MA, Kawate N, Kubo Y, Pathirana IN, Büllesbach EE, Hatoya S, Inaba T, Takahashi M, Tamada H. Hannan MA, et al. Theriogenology. 2015 Oct 15;84(7):1176-84. doi: 10.1016/j.theriogenology.2015.06.021. Epub 2015 Jul 2. Theriogenology. 2015. PMID: 26220663 - INSL3/RXFP2 signaling in testicular descent.
Feng S, Ferlin A, Truong A, Bathgate R, Wade JD, Corbett S, Han S, Tannour-Louet M, Lamb DJ, Foresta C, Agoulnik AI. Feng S, et al. Ann N Y Acad Sci. 2009 Apr;1160:197-204. doi: 10.1111/j.1749-6632.2009.03841.x. Ann N Y Acad Sci. 2009. PMID: 19416188 Free PMC article. Review. - Factors controlling testis descent.
Hughes IA, Acerini CL. Hughes IA, et al. Eur J Endocrinol. 2008 Dec;159 Suppl 1:S75-82. doi: 10.1530/EJE-08-0458. Epub 2008 Jul 22. Eur J Endocrinol. 2008. PMID: 18647820 Review.
Cited by
- Recapitulating Evolutionary Divergence in a Single Cis-Regulatory Element Is Sufficient to Cause Expression Changes of the Lens Gene Tdrd7.
Roscito JG, Subramanian K, Naumann R, Sarov M, Shevchenko A, Bogdanova A, Kurth T, Foerster L, Kreysing M, Hiller M. Roscito JG, et al. Mol Biol Evol. 2021 Jan 23;38(2):380-392. doi: 10.1093/molbev/msaa212. Mol Biol Evol. 2021. PMID: 32853335 Free PMC article. - Therian origin of INSL3/RXFP2-driven testicular descent in mammals.
Menzies BR, Tarulli GA, Frankenberg SR, Pask AJ. Menzies BR, et al. Front Cell Dev Biol. 2024 Feb 2;12:1353598. doi: 10.3389/fcell.2024.1353598. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38380341 Free PMC article. - Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains.
Jebb D, Hiller M. Jebb D, et al. Elife. 2018 Oct 16;7:e38906. doi: 10.7554/eLife.38906. Elife. 2018. PMID: 30322448 Free PMC article. - Age cohort definition and evidence of sexual dimorphism in the Southern Hairy-nosed Wombat (Lasiorhinus latifrons), a large Australian marsupial.
Kleemann SL, Taggart DA. Kleemann SL, et al. J Mammal. 2025 Feb 22;106(3):702-711. doi: 10.1093/jmammal/gyaf009. eCollection 2025 Jun. J Mammal. 2025. PMID: 40520352 Free PMC article. - The fourspine stickleback (Apeltes quadracus) has an XY sex chromosome system with polymorphic inversions on both X and Y chromosomes.
Liu Z, Herbert AL, Chan YF, Kučka M, Kingsley DM, Peichel CL. Liu Z, et al. PLoS Genet. 2025 May 9;21(5):e1011465. doi: 10.1371/journal.pgen.1011465. eCollection 2025 May. PLoS Genet. 2025. PMID: 40344089 Free PMC article.
References
- Asher RJ, Helgen KM. Nomenclature and placental mammal phylogeny. BMC Evol Biol. 2010;10:102 doi: 10.1186/1471-2148-10-102 . - DOI - PMC - PubMed
- Werdelin L, Nilsonne A. The evolution of the scrotum and testicular descent in mammals: a phylogenetic view. Journal of theoretical biology. 1999;196(1):61–72. doi: 10.1006/jtbi.1998.0821 . - DOI - PubMed
- Kleisner K, Ivell R, Flegr J. The evolutionary history of testicular externalization and the origin of the scrotum. Journal of biosciences. 2010;35(1):27–37. . - PubMed
- Rommel SA, Pabst DA, McLellan WA, Mead JG, Potter CW. Anatomical evidence for a countercurrent heat exchanger associated with dolphin testes. The Anatomical record. 1992;232(1):150–6. doi: 10.1002/ar.1092320117 . - DOI - PubMed
- Rommel SA, Early GA, Matassa KA, Pabst DA, McLellan WA. Venous structures associated with thermoregulation of phocid seal reproductive organs. The Anatomical record. 1995;243(3):390–402. doi: 10.1002/ar.1092430314 . - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical