Fusobacterium Genomics Using MinION and Illumina Sequencing Enables Genome Completion and Correction - PubMed (original) (raw)
Fusobacterium Genomics Using MinION and Illumina Sequencing Enables Genome Completion and Correction
S Michelle Todd et al. mSphere. 2018.
Abstract
Understanding the virulence mechanisms of human pathogens from the genus Fusobacterium has been hindered by a lack of properly assembled and annotated genomes. Here we report the first complete genomes for seven Fusobacterium strains, as well as resequencing of the reference strain Fusobacterium nucleatum subsp. nucleatum ATCC 25586 (total of seven species; total of eight genomes). A highly efficient and cost-effective sequencing pipeline was achieved using sample multiplexing for short-read Illumina (150 bp) and long-read Oxford Nanopore MinION (>80 kbp) platforms, coupled with genome assembly using the open-source software Unicycler. Compared to currently available draft assemblies (previously 24 to 67 contigs), these genomes are highly accurate and consist of only one complete chromosome. We present the complete genome sequence of F. nucleatum subsp. nucleatum ATCC 23726, a genetically tractable and biomedically important strain and, in addition, reveal that the previous F. nucleatum subsp. nucleatum ATCC 25586 genome assembly contains a 452-kb genomic inversion that has been corrected using our sequencing and assembly pipeline. To enable genomic analyses by the scientific community, we concurrently used these genomes to launch FusoPortal, a repository of interactive and downloadable genomic data, genome maps, gene annotations, and protein functional analyses and classifications. In summary, this report provides detailed methods for accurately sequencing, assembling, and annotating Fusobacterium genomes, while focusing on using open-source software to foster the availability of reproducible and open data. This resource will enhance efforts to properly identify virulence proteins that may contribute to a repertoire of diseases that includes periodontitis, preterm birth, and colorectal cancer.IMPORTANCEFusobacterium spp. are Gram-negative, oral bacteria that are increasingly associated with human pathologies as diverse as periodontitis, preterm birth, and colorectal cancer. While a recent surge in F. nucleatum research has increased our understanding of this human pathogen, a lack of complete genomes has hindered the identification and characterization of associated host-pathogen virulence factors. Here we report the first eight complete Fusobacterium genomes sequenced using an Oxford Nanopore MinION and Illumina sequencing pipeline and assembled using the open-source program Unicycler. These genomes are highly accurate, and seven of the genomes represent the first complete sequences for each strain. In summary, the FusoPortal resource provides a publicly available resource that will guide future genetic, bioinformatic, and biochemical experiments to characterize this genus of emerging human pathogens.
Keywords: Fusobacterium; Fusobacterium nucleatum; Illumina; MinION; cancer; colorectal cancer.
Copyright © 2018 Todd et al.
Figures
FIG 1
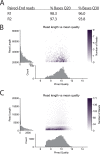
Phred analysis of Illumina and MinION reads for the F. nucleatum subsp. nucleatum ATCC 25586 genome. (A) Q scores for Illumina reads. (B) Q scores (Phred quality) for MinION reads 20 to 80 kb in length. (C) Q scores for MinION reads up to 20 kb in length.
FIG 2
Statistics and mapping of F. necrophorum_funduliforme_ 1_1_36S MinION long reads. After complete genome assembly, MinION reads were mapped to the F. necrophorum_funduliforme_ 1_1_36S genome using Geneious version 9.1.4 software.
FIG 3
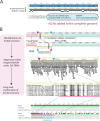
Genome assembly and annotation of eight Fusobacterium genomes from seven species. Short-read-only and complete genome assembly representations were created using Bandage (21).
FIG 4
Analysis of Fusobacterium nucleatum ATCC 23726 genomes compared to previous builds. (A) Alignment of the complete F. nucleatum subsp. nucleatum ATCC 23726 genome with the 67-contig draft assembly (GenBank accession number
ADVK01000000
). (B) Confirmation of a 452-kb genomic inversion in the previous F. nucleatum subsp. nucleatum ATCC 25586 genome assembly (GenBank accession number
GCA_000007325.1
).
Similar articles
- FusoPortal: an Interactive Repository of Hybrid MinION-Sequenced Fusobacterium Genomes Improves Gene Identification and Characterization.
Sanders BE, Umana A, Lemkul JA, Slade DJ. Sanders BE, et al. mSphere. 2018 Jul 5;3(4):e00228-18. doi: 10.1128/mSphere.00228-18. mSphere. 2018. PMID: 29976644 Free PMC article. - Utilizing Whole Fusobacterium Genomes To Identify, Correct, and Characterize Potential Virulence Protein Families.
Umaña A, Sanders BE, Yoo CC, Casasanta MA, Udayasuryan B, Verbridge SS, Slade DJ. Umaña A, et al. J Bacteriol. 2019 Nov 5;201(23):e00273-19. doi: 10.1128/JB.00273-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501282 Free PMC article. - Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium.
Karpathy SE, Qin X, Gioia J, Jiang H, Liu Y, Petrosino JF, Yerrapragada S, Fox GE, Haake SK, Weinstock GM, Highlander SK. Karpathy SE, et al. PLoS One. 2007 Aug 1;2(7):e659. doi: 10.1371/journal.pone.0000659. PLoS One. 2007. PMID: 17668047 Free PMC article. - Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer.
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Nosho K, et al. World J Gastroenterol. 2016 Jan 14;22(2):557-66. doi: 10.3748/wjg.v22.i2.557. World J Gastroenterol. 2016. PMID: 26811607 Free PMC article. Review. - Fusobacterium nucleatum: an emerging gut pathogen?
Allen-Vercoe E, Strauss J, Chadee K. Allen-Vercoe E, et al. Gut Microbes. 2011 Sep 1;2(5):294-8. doi: 10.4161/gmic.2.5.18603. Epub 2011 Sep 1. Gut Microbes. 2011. PMID: 22067936 Review.
Cited by
- Genome Sequences for Two Acinetobacter baumannii Strains Obtained Using the Unicycler Hybrid Assembly Pipeline.
Farrow JM 3rd, Pesci EC, Slade DJ. Farrow JM 3rd, et al. Microbiol Resour Announc. 2021 Mar 11;10(10):e00017-21. doi: 10.1128/MRA.00017-21. Microbiol Resour Announc. 2021. PMID: 33707317 Free PMC article. - Fusobacterium nucleatum - symbiont, opportunist and oncobacterium.
Brennan CA, Garrett WS. Brennan CA, et al. Nat Rev Microbiol. 2019 Mar;17(3):156-166. doi: 10.1038/s41579-018-0129-6. Nat Rev Microbiol. 2019. PMID: 30546113 Free PMC article. Review. - GenoFig: a user-friendly application for the visualization and comparison of genomic regions.
Branger M, Leclercq SO. Branger M, et al. Bioinformatics. 2024 Jun 3;40(6):btae372. doi: 10.1093/bioinformatics/btae372. Bioinformatics. 2024. PMID: 38870520 Free PMC article. - Tumour microbiomes and Fusobacterium genomics in Vietnamese colorectal cancer patients.
Tran HNH, Thu TNH, Nguyen PH, Vo CN, Doan KV, Nguyen Ngoc Minh C, Nguyen NT, Ta VND, Vu KA, Hua TD, Nguyen TNT, Van TT, Pham Duc T, Duong BL, Nguyen PM, Hoang VC, Pham DT, Thwaites GE, Hall LJ, Slade DJ, Baker S, Tran VH, Chung The H. Tran HNH, et al. NPJ Biofilms Microbiomes. 2022 Oct 29;8(1):87. doi: 10.1038/s41522-022-00351-7. NPJ Biofilms Microbiomes. 2022. PMID: 36307484 Free PMC article. - Completing Circular Bacterial Genomes With Assembly Complexity by Using a Sampling Strategy From a Single MinION Run With Barcoding.
Liao YC, Cheng HW, Wu HC, Kuo SC, Lauderdale TY, Chen FJ. Liao YC, et al. Front Microbiol. 2019 Sep 4;10:2068. doi: 10.3389/fmicb.2019.02068. eCollection 2019. Front Microbiol. 2019. PMID: 31551994 Free PMC article.
References
- Signat B, Roques C, Poulet P, Duffaut D. 2011. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 13:25–36. - PubMed
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. doi:10.1101/gr.126573.111. - DOI - PMC - PubMed
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi:10.1016/j.chom.2013.07.007. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous