Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer - PubMed (original) (raw)
Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer
Zsofia Sztupinszki et al. NPJ Breast Cancer. 2018.
Abstract
The first genomic scar-based homologous recombination deficiency (HRD) measures were produced using SNP arrays. As array-based technology has been largely replaced by next generation sequencing approaches, it has become important to develop algorithms that derive the same type of genomic scar scores from next generation sequencing (whole exome "WXS", whole genome "WGS") data. In order to perform this analysis, we introduce here the scarHRD R package and show that using this method the SNP array-based and next generation sequencing-based derivation of HRD scores show good correlation (Pearson correlation between 0.73 and 0.87 depending on the actual HRD measure) and that the NGS-based HRD scores distinguish similarly well between BRCA mutant and BRCA wild-type cases in a cohort of triple-negative breast cancer patients of the TCGA data set.
Conflict of interest statement
N.J.B., A.C.E., and Zo.S are listed as co-inventors on a patent on telomeric allelic imbalance, which is owned by Children’s Hospital Boston and licensed to Myriad Genetics. The remaining authors declare no competing interests.
Figures
Fig. 1
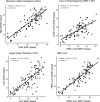
Correlation between Affymetrix SNP 6.0 array-based and whole exome sequencing-based measurements of homologous recombination deficiency (telomeric allelic imbalance, loss of heterozygosity, large-scale transitions, and the sum of these estimates)
Fig. 2
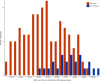
Distribution of HRD-sum values in BRCA1/2 deficient and in BRCA1/2 intact triple-negative breast cancer samples from TCGA. HRD-sum values were determined with the scarHRD R package
Similar articles
- Using whole-genome sequencing data to derive the homologous recombination deficiency scores.
de Luca XM, Newell F, Kazakoff SH, Hartel G, McCart Reed AE, Holmes O, Xu Q, Wood S, Leonard C, Pearson JV, Lakhani SR, Waddell N, Nones K, Simpson PT. de Luca XM, et al. NPJ Breast Cancer. 2020 Aug 7;6:33. doi: 10.1038/s41523-020-0172-0. eCollection 2020. NPJ Breast Cancer. 2020. PMID: 32818150 Free PMC article. - Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes.
Timms KM, Abkevich V, Hughes E, Neff C, Reid J, Morris B, Kalva S, Potter J, Tran TV, Chen J, Iliev D, Sangale Z, Tikishvili E, Perry M, Zharkikh A, Gutin A, Lanchbury JS. Timms KM, et al. Breast Cancer Res. 2014 Dec 5;16(6):475. doi: 10.1186/s13058-014-0475-x. Breast Cancer Res. 2014. PMID: 25475740 Free PMC article. - Concordance between single-nucleotide polymorphism-based genomic instability assays and a next-generation sequencing-based homologous recombination deficiency test.
Cristescu R, Liu XQ, Arreaza G, Chen C, Albright A, Qiu P, Marton MJ. Cristescu R, et al. BMC Cancer. 2022 Dec 14;22(1):1310. doi: 10.1186/s12885-022-10197-z. BMC Cancer. 2022. PMID: 36517748 Free PMC article. - Biomarkers for Homologous Recombination Deficiency in Cancer.
Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Hoppe MM, et al. J Natl Cancer Inst. 2018 Jul 1;110(7):704-713. doi: 10.1093/jnci/djy085. J Natl Cancer Inst. 2018. PMID: 29788099 Review. - Using SAAS-CNV to Detect and Characterize Somatic Copy Number Alterations in Cancer Genomes from Next Generation Sequencing and SNP Array Data.
Zhang Z, Hao K. Zhang Z, et al. Methods Mol Biol. 2018;1833:29-47. doi: 10.1007/978-1-4939-8666-8_2. Methods Mol Biol. 2018. PMID: 30039361 Review.
Cited by
- Direct prediction of Homologous Recombination Deficiency from routine histology in ten different tumor types with attention-based Multiple Instance Learning: a development and validation study.
Loeffler CML, El Nahhas OSM, Muti HS, Seibel T, Cifci D, van Treeck M, Gustav M, Carrero ZI, Gaisa NT, Lehmann KV, Leary A, Selenica P, Reis-Filho JS, Bruechle NO, Kather JN. Loeffler CML, et al. medRxiv [Preprint]. 2023 Mar 10:2023.03.08.23286975. doi: 10.1101/2023.03.08.23286975. medRxiv. 2023. PMID: 36945540 Free PMC article. Updated. Preprint. - Utility of exome sequencing in routine care for metastatic colorectal cancer.
D'Agay MG, Galland L, Tharin Z, Truntzer C, Ghiringhelli F. D'Agay MG, et al. Mol Clin Oncol. 2021 Nov;15(5):229. doi: 10.3892/mco.2021.2392. Epub 2021 Sep 10. Mol Clin Oncol. 2021. PMID: 34631054 Free PMC article. - Molecular landscape and therapeutic alterations in Asian soft-tissue sarcoma patients.
Gan M, Zhang C, Qiu L, Wang Y, Bao H, Yu R, Liu R, Wu X, Shao Y, Hou P, Fei Z. Gan M, et al. Cancer Med. 2022 Nov;11(21):4070-4078. doi: 10.1002/cam4.4725. Epub 2022 May 18. Cancer Med. 2022. PMID: 35586877 Free PMC article. - Aberrant Transcript Usage Is Associated with Homologous Recombination Deficiency and Predicts Therapeutic Response.
Kang HG, Hwangbo H, Kim MJ, Kim S, Lee EJ, Park MJ, Kim JW, Kim BG, Cho EH, Chang S, Lee JY, Choi JK. Kang HG, et al. Cancer Res. 2022 Jan 1;82(1):142-154. doi: 10.1158/0008-5472.CAN-21-2023. Epub 2021 Oct 28. Cancer Res. 2022. PMID: 34711610 Free PMC article. - Multi-Gene Testing Overview with a Clinical Perspective in Metastatic Triple-Negative Breast Cancer.
Dameri M, Ferrando L, Cirmena G, Vernieri C, Pruneri G, Ballestrero A, Zoppoli G. Dameri M, et al. Int J Mol Sci. 2021 Jul 1;22(13):7154. doi: 10.3390/ijms22137154. Int J Mol Sci. 2021. PMID: 34281208 Free PMC article. Review.
References
- Birkbak NJ, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials