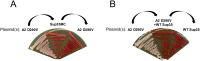Sequence features governing aggregation or degradation of prion-like proteins - PubMed (original) (raw)
Sequence features governing aggregation or degradation of prion-like proteins
Sean M Cascarina et al. PLoS Genet. 2018.
Abstract
Enhanced protein aggregation and/or impaired clearance of aggregates can lead to neurodegenerative disorders such as Alzheimer's Disease, Huntington's Disease, and prion diseases. Therefore, many protein quality control factors specialize in recognizing and degrading aggregation-prone proteins. Prions, which generally result from self-propagating protein aggregates, must therefore evade or outcompete these quality control systems in order to form and propagate in a cellular context. We developed a genetic screen in yeast that allowed us to explore the sequence features that promote degradation versus aggregation of a model glutamine/asparagine (Q/N)-rich prion domain from the yeast prion protein, Sup35, and two model glycine (G)-rich prion-like domains from the human proteins hnRNPA1 and hnRNPA2. Unexpectedly, we found that aggregation propensity and degradation propensity could be uncoupled in multiple ways. First, only a subset of classically aggregation-promoting amino acids elicited a strong degradation response in the G-rich prion-like domains. Specifically, large aliphatic residues enhanced degradation of the prion-like domains, whereas aromatic residues promoted prion aggregation without enhancing degradation. Second, the degradation-promoting effect of aliphatic residues was suppressed in the context of the Q/N-rich prion domain, and instead led to a dose-dependent increase in the frequency of spontaneous prion formation. Degradation suppression correlated with Q/N content of the surrounding prion domain, potentially indicating an underappreciated activity for these residues in yeast prion domains. Collectively, these results provide key insights into how certain aggregation-prone proteins may evade protein quality control degradation systems.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Fig 1. The A2-Sup35 protein exhibits PrLD-dependent prion activity.
(A) A2-Sup35 prion maintenance requires continual expression of the A2-Sup35 prion domain. A covering plasmid expressing a copy of Sup35 lacking the prion domain was shuffled into a [PRION+] isolate (A2 D290V). In the [PRION+] strain, cells became _ade_- upon loss of the A2-Sup35 plasmid, and remained _ade_- when the plasmid was re-introduced. (B) A2-Sup35 prions are unable to convert wild-type Sup35. Co-expression of wild-type Sup35 in the [PRION+] strain resulted in a reversion to the _ade_- phenotype that was maintained upon loss of the A2-Sup35 plasmid, suggesting that the A2-Sup35 prion form is not efficiently transmitted to wild-type Sup35.
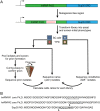
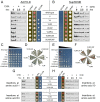
Fig 2. Mutagenesis method.
(A) The Sup35 prion domain contains an N-terminal Q/N-rich prion nucleation domain, and an oligopeptide repeat domain. The nucleation domain in full-length Sup35 was replaced with the core PrLDs from hnRNPA1 and hnRNPA2. An 8-amino-acid segment in each of the fusion proteins was then randomly mutagenized. Mutants were expressed as the sole copy of Sup35 in the cell. Library members were screened for their initial adenine phenotype, and for the ability to form prions. (B) Sequences of the core PrLDs from hnRNPA1 and A2, and the nucleation domain of Sup35. The underlined segments of hnRNPA1 and A2 were mutagenized in this study, while the underlined segment of Sup35 was mutagenized previously [44]. Arrow heads indicate the sites of hydrophobic insertion in Figs 7A and 6B.
Fig 3. Similar amino acid biases govern the ADE + phenotype within the A1 and A2 PrLD’s.
Comparison of the log-odds ratio for each amino acid between the A1 and A2 libraries. The log-odds ratios reflects the degree of over-/under-representation of each amino acid among constitutive ADE + isolates, relative to ade - isolates. Colors correspond to the amino acid groups in Table 1.
Fig 4. Hydrophobic peptides promote degradation of the A2 PrLD but not the Sup35 nucleation domain.
(A) The ADE + phenotype for A2-Sup35 mutants is associated with increased protein turnover. ADE + and ade - isolates expressing the indicated A2-Sup35 fusion as the sole copy of Sup35 in the cell were plated on SC-ade and YPD to confirm phenotypes originally observed in the mutagenesis/screening method. Protein levels were assessed by western blot after treatment with CHX. The ADE + phenotype is associated with accelerated degradation of the fusion protein. Wild-type sequences are the respective sequences from wild-type Sup35 and the A2 PrLD. (B) In the context of the Sup35 ND, all peptides conferred an ade - phenotype and did not accelerate degradation. Western blots were quantified for all A2 PrLD (C) and Sup35 ND (D) mutants. Data represent means ± SDs (n≥3).
Fig 5. Degradation of A2 PrLDs occurs via the ubiquitin-proteasome system.
Addition of MG-132 (+) 1hr prior to the addition of CHX prevents degradation of A2-Sup35 fusions (SGNYNDFG is the sequence in the corresponding region of the wild-type A2 PrLD).
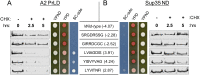
Fig 6. Amino acid degradation scores are sufficient to identify degradation-promoting and inhibiting peptides from an independent peptide library.
(A) Predicted degradation-promoting (LVIAGDIS, YISVYVAG, and LYVITNFI) or inhibiting (SRGDRSSG and GIRRDCGC) peptides were substituted into the A2 PrLD (numbers in parentheses indicate the sum of the individual amino acid scores derived from the A2 PrLD degradation library). Predicted degradation-promoting peptides led to an ADE + phenotype and accelerated degradation of the A2 PrLD. Predicted degradation-inhibiting peptides led to the ade - and showed no increase in degradation rate of the A2-Sup35 fusion. (B) In the context of the Sup35 ND, all peptides were stable over 5 hrs and conferred a predominantly ade - phenotype.
Fig 7. Degradation of the A2 PrLD and prion aggregation of Sup35 occur at similar hydrophobic content thresholds.
(A) Two or more hydrophobic residues inserted into the A2 PrLD resulted in a robust ADE + phenotype and accelerated degradation of the A2 PrLD. (B) Insertion of three or more hydrophobic residues into Sup35 led to a progressive increase in the frequency of white sectors on YPD without affecting Sup35 turnover. (C) To quantify the frequency of ADE + colony formation, serial dilutions of cells expressing each A2-Sup35 fusion were plated onto SC-ade. Degradation of the A2 PrLD upon insertion of two or more hydrophobic residues was correlated with a binary-like switch from _ade_- to ADE+. (D) ADE+ isolates from the A2 PrLD mutants were not curable by GuHCl. To test for curability of the ADE+ phenotype, individual ADE+ colonies were streaked on YPD (-) or YPD plus 4mM GuHCl (+), and then re-streaked onto YPD to test for loss of the ADE+ phenotype. (E) Insertion of three or more hydrophobic residues in the Sup35 ND leads to a progressive increase in ADE+ growth. (F) ADE+ isolates from the Sup35 mutants were curable by GuHCl, consistent with the ADE + phenotype resulting from prion formation. (G) Insertion of hydrophobic amino acids at other positions in the A2 PrLD also promoted protein degradation. (H) Insertion of hydrophobic amino acids at other positions in the Sup35 nucleation domain had little or no effect on protein turnover.
Fig 8. Degradation of the A2 PrLD and stability of the Sup35 ND upon insertion of hydrophobic residues do not depend on the Sup35 oligopeptide repeat domain (ORD), M-domain, or C-domain.
Progressively increasing hydrophobic content in the A2 PrLD (A) when fused to GFP alone (top) or with the remainder of the Sup35NM domains and tandem FLAG tags (bottom) enhances degradation rate of the A2 PrLD fusion proteins. By contrast, insertion of hydrophobic residues in the Sup35 ND (B) fused to GFP (top) or Sup35NM-2xFLAG (bottom) does not decrease stability of the Sup35 ND fusion proteins. Data represent means ± SDs (n = 3).
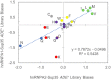
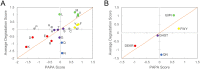
Fig 9. Yeast prion domains are enriched in amino acids that are prion-prone but not degradation-promoting.
Average degradation scores from the A1 PrLD and A2 PrLD libraries are plotted against yeast prion propensity scores for individual amino acids (A) or amino acid groups (B). Within native yeast prion domains, commonly occurring amino acids (Q, N, and aromatic amino acids) exhibit a combination of high prion propensity and low degradation propensity. Colors correspond to the amino acid groups in Table 1 and Fig 3.
Fig 10. High Q/N content of Sup35 antagonizes degradation.
(A) Sequences of the partial or full Q/N/G substitution constructs. Glycines are indicated in red, while glutamines and asparagines are in green. (B) Partial or full substitution of Q/N residues for G within the Sup35 nucleation domain resulted in the ADE + degradation phenotype (left), and a step-wise increase in degradation rate (right). Partial substitution of G residues for Q/N residues within the A2 PrLD resulted in an ade - phenotype (left), and corresponding decrease in degradation rate (right). Full substitution of remaining G residues for QN residues had mixed effects.
Similar articles
- The effects of glutamine/asparagine content on aggregation and heterologous prion induction by yeast prion-like domains.
Shattuck JE, Waechter AC, Ross ED. Shattuck JE, et al. Prion. 2017 Jul 4;11(4):249-264. doi: 10.1080/19336896.2017.1344806. Epub 2017 Jun 30. Prion. 2017. PMID: 28665753 Free PMC article. - Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro.
Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Derkatch IL, et al. Proc Natl Acad Sci U S A. 2004 Aug 31;101(35):12934-9. doi: 10.1073/pnas.0404968101. Epub 2004 Aug 23. Proc Natl Acad Sci U S A. 2004. PMID: 15326312 Free PMC article. - Increasing prion propensity by hydrophobic insertion.
Gonzalez Nelson AC, Paul KR, Petri M, Flores N, Rogge RA, Cascarina SM, Ross ED. Gonzalez Nelson AC, et al. PLoS One. 2014 Feb 20;9(2):e89286. doi: 10.1371/journal.pone.0089286. eCollection 2014. PLoS One. 2014. PMID: 24586661 Free PMC article. - Aggregation and degradation scales for prion-like domains: sequence features and context weigh in.
Cascarina SM, Ross ED. Cascarina SM, et al. Curr Genet. 2019 Apr;65(2):387-392. doi: 10.1007/s00294-018-0890-0. Epub 2018 Oct 11. Curr Genet. 2019. PMID: 30310993 Review. - Fused in Sarcoma: Properties, Self-Assembly and Correlation with Neurodegenerative Diseases.
Chen C, Ding X, Akram N, Xue S, Luo SZ. Chen C, et al. Molecules. 2019 Apr 24;24(8):1622. doi: 10.3390/molecules24081622. Molecules. 2019. PMID: 31022909 Free PMC article. Review.
Cited by
- Variable absorption of mutational trends by prion-forming domains during Saccharomycetes evolution.
Harrison PM. Harrison PM. PeerJ. 2020 Aug 6;8:e9669. doi: 10.7717/peerj.9669. eCollection 2020. PeerJ. 2020. PMID: 32844065 Free PMC article. - Hsp40/JDP Requirements for the Propagation of Synthetic Yeast Prions.
Miller SC, Wegrzynowicz AK, Cole SJ, Hayward RE, Ganser SJ, Hines JK. Miller SC, et al. Viruses. 2022 Sep 30;14(10):2160. doi: 10.3390/v14102160. Viruses. 2022. PMID: 36298715 Free PMC article. - Sky1: at the intersection of prion-like proteins and stress granule regulation.
Shattuck JE, Cascarina SM, Paul KR, Ross ED. Shattuck JE, et al. Curr Genet. 2020 Jun;66(3):463-468. doi: 10.1007/s00294-019-01044-z. Epub 2019 Nov 19. Curr Genet. 2020. PMID: 31745569 Free PMC article. Review. - Yeast Models of Prion-Like Proteins That Cause Amyotrophic Lateral Sclerosis Reveal Pathogenic Mechanisms.
Monahan ZT, Rhoads SN, Yee DS, Shewmaker FP. Monahan ZT, et al. Front Mol Neurosci. 2018 Dec 11;11:453. doi: 10.3389/fnmol.2018.00453. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30618605 Free PMC article. Review. - The molecular language of membraneless organelles.
Gomes E, Shorter J. Gomes E, et al. J Biol Chem. 2019 May 3;294(18):7115-7127. doi: 10.1074/jbc.TM118.001192. Epub 2018 Jul 25. J Biol Chem. 2019. PMID: 30045872 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases