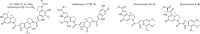A radical S-adenosyl-L-methionine enzyme and a methyltransferase catalyze cyclopropane formation in natural product biosynthesis - PubMed (original) (raw)
A radical S-adenosyl-L-methionine enzyme and a methyltransferase catalyze cyclopropane formation in natural product biosynthesis
Wen-Bing Jin et al. Nat Commun. 2018.
Abstract
Cyclopropanation of unactivated olefinic bonds via addition of a reactive one-carbon species is well developed in synthetic chemistry, whereas natural cyclopropane biosynthesis employing this strategy is very limited. Here, we identify a two-component cyclopropanase system, composed of a HemN-like radical S-adenosyl-L-methionine (SAM) enzyme C10P and a methyltransferase C10Q, catalyzes chemically challenging cyclopropanation in the antitumor antibiotic CC-1065 biosynthesis. C10P uses its [4Fe-4S] cluster for reductive cleavage of the first SAM to yield a highly reactive 5'-deoxyadenosyl radical, which abstracts a hydrogen from the second SAM to produce a SAM methylene radical that adds to an sp2-hybridized carbon of substrate to form a SAM-substrate adduct. C10Q converts this adduct to CC-1065 via an intramolecular SN2 cyclization mechanism with elimination of S-adenosylhomocysteine. This cyclopropanation strategy not only expands the enzymatic reactions catalyzed by the radical SAM enzymes and methyltransferases, but also sheds light on previously unnoticed aspects of the versatile SAM-based biochemistry.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
Chemical structures of the spirocyclopropylcyclohexadienone family of natural products. This family of natural products includes CC-1065 (1), gilvusmycin (2), yatakemycin (YTM, 3), duocarmycin SA (4), and duocarmycin A (5)
Fig. 2
Characterization of a two-component cyclopropanase system. a–h HPLC analysis of the relevant metabolites (UV at 374 nm). a Standard of 1.b Standard of 6. c Wild type Streptomyces zelensis NRRL 11183. d The Δ_c10P_ mutant. e The Δ_c10Q_ mutant. f The Δ_c10Q_ mutant complementated with the c10Q gene. g The Δ_c10P_ mutant complementated with the Swoo_2002 gene. h The Δ_c10P_ mutant complementated with the c10P gene. i Structures of 6 and 7. j–q HPLC analysis of enzymatic products. j–m are the control reactions without C10P, C10Q, Na2S2O4, and SAM, respectively. n Standard of 1. o The complete reaction for C10P and C10Q. p The complete reaction for Swoo_2002 and C10Q. q The reaction for Swoo_2002 and C10Q H138A. All the in vitro enzymatic activity assays were performed in an anaerobic glove box with less than 1 ppm of O2. Reactions were conducted in Tris•HCl buffer (Tris 50 mM, NaCl 100 mM, glycerol 10%, pH 8.0) with the following composition: 1 μM reconstituted Swoo_2002, 2 μM C10Q, 10 μM substrate 6, 1 mM SAM, 5 mM DTT, 5 mM MgCl2, 5 mM Na2S2O4 and 7% DMSO. The reactions were incubated at 28 °C for 12 h
Fig. 3
Analysis of reaction products from the cyclopropanation process. a–e HPLC analysis of the enzymatic products (UV at 260 nm). a Standard of 5′-dA. b Standard of SAH. c The control reaction using boiling-inactivated enzymes. d, e are the concentrated products from a large-scale enzymatic assay terminated at 4 h and subjected to evaporation and lyophilization, respectively. f Time-course analysis of the concentration changes of 1, 6, 7, 5′-dA, and SAH from the cyclopropanation process. g, h are the HR-MS analyses of the intermediate 8 from the reactions using SAM and CD3-SAM, respectively. i MS/MS analysis of 8 using SAM. j The chemical structure and calculated molecular weight of the intermediate 8
Fig. 4
Isotope labeling investigations into the cyclopropanation process. a–f HR-MS analysis of the products from reactions using SAM or CD3-SAM. a, c, e are the products of 5′-dA, 1, and 7, respectively, from the assay using SAM. b, d, f are the products of D-5′-dA, D 2 -1, and D 2 -7, respectively, from the assay using CD3-SAM. g-j HR-MS analysis of the products from reactions using H2O or D2O. g, i are the products of 1 and 7, respectively, from the assay using H2O. h, j are the products of D-1 and D-7, respectively, from the assay using D2O. k and l are the NMR analyses of the enzymatic product 7 from H2O (7, 1H-NMR) and D2O (D-7, 2H-NMR)
Fig. 5
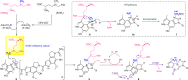
Proposed mechanism of the cyclopropanation process. Upon dithionite reduction, the [4Fe-4S]2+ from the HemN-like radical SAM enzyme is converted to [4Fe-4S]+, which triggers the reductive cleavage of the first molecule of SAM1 to yield a highly reactive dAdo radical. Then, the dAdo radical abstracts a hydrogen atom from the methyl group of the second molecule of SAM2. A SAM methylene radical is thus produced and then adds to the C-11 position of the substrate 6 to generate a radical intermediate 9. The carbon-centered radical at C-12 in 9 abstracts a solvent-exchangeable proton to produce the intermediate 8. Subsequently, the His-138 residue from C10Q likely functions as a critical base and deprotonates the phenolic hydroxyl group (C-6) of 8, which induces the intramolecular SN2 cyclopropanation to yield 1 with elimination of SAH as a co-product. On the other hand, the intermediate 8 may be non-enzymatically converted to the intermediate 10 containing an exocyclic double bond via release of SAH, followed by rapid and thermodynamic driving isomerization to give a methylated off-pathway compound 7
Similar articles
- Crystallographic snapshots of sulfur insertion by lipoyl synthase.
McLaughlin MI, Lanz ND, Goldman PJ, Lee KH, Booker SJ, Drennan CL. McLaughlin MI, et al. Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):9446-50. doi: 10.1073/pnas.1602486113. Epub 2016 Aug 9. Proc Natl Acad Sci U S A. 2016. PMID: 27506792 Free PMC article. - Cfr and RlmN contain a single [4Fe-4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: methyl transfer by SN2 displacement and radical generation.
Grove TL, Radle MI, Krebs C, Booker SJ. Grove TL, et al. J Am Chem Soc. 2011 Dec 14;133(49):19586-9. doi: 10.1021/ja207327v. Epub 2011 Nov 18. J Am Chem Soc. 2011. PMID: 21916495 Free PMC article. - Auxiliary iron-sulfur cofactors in radical SAM enzymes.
Lanz ND, Booker SJ. Lanz ND, et al. Biochim Biophys Acta. 2015 Jun;1853(6):1316-34. doi: 10.1016/j.bbamcr.2015.01.002. Epub 2015 Jan 15. Biochim Biophys Acta. 2015. PMID: 25597998 Review. - _C_-Methylation of _S_-adenosyl-L-Methionine Occurs Prior to Cyclopropanation in the Biosynthesis of 1-Amino-2-Methylcyclopropanecarboxylic Acid (Norcoronamic Acid) in a Bacterium.
Maruyama C, Chinone Y, Sato S, Kudo F, Ohsawa K, Kubota J, Hashimoto J, Kozone I, Doi T, Shin-Ya K, Eguchi T, Hamano Y. Maruyama C, et al. Biomolecules. 2020 May 16;10(5):775. doi: 10.3390/biom10050775. Biomolecules. 2020. PMID: 32429436 Free PMC article. - Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes.
Lanz ND, Booker SJ. Lanz ND, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1196-212. doi: 10.1016/j.bbapap.2012.07.009. Epub 2012 Jul 28. Biochim Biophys Acta. 2012. PMID: 22846545 Review.
Cited by
- Astrocytic ALKBH5 in stress response contributes to depressive-like behaviors in mice.
Guo F, Fan J, Liu JM, Kong PL, Ren J, Mo JW, Lu CL, Zhong QL, Chen LY, Jiang HT, Zhang C, Wen YL, Gu TT, Li SJ, Fang YY, Pan BX, Gao TM, Cao X. Guo F, et al. Nat Commun. 2024 May 21;15(1):4347. doi: 10.1038/s41467-024-48730-2. Nat Commun. 2024. PMID: 38773146 Free PMC article. - Diversity of the reaction mechanisms of SAM-dependent enzymes.
Sun Q, Huang M, Wei Y. Sun Q, et al. Acta Pharm Sin B. 2021 Mar;11(3):632-650. doi: 10.1016/j.apsb.2020.08.011. Epub 2020 Aug 26. Acta Pharm Sin B. 2021. PMID: 33777672 Free PMC article. Review. - Functional Diversity of HemN-like Proteins.
Cheng J, Liu WQ, Zhu X, Zhang Q. Cheng J, et al. ACS Bio Med Chem Au. 2022 Jan 18;2(2):109-119. doi: 10.1021/acsbiomedchemau.1c00058. eCollection 2022 Apr 20. ACS Bio Med Chem Au. 2022. PMID: 37101745 Free PMC article. Review. - Biosynthesis of coelulatin for the methylation of anthraquinone featuring HemN-like radical S-adenosyl-L-methionine enzyme.
Nie L, Wei T, Cao M, Lyu Y, Wang S, Feng Z. Nie L, et al. Front Microbiol. 2022 Nov 17;13:1040900. doi: 10.3389/fmicb.2022.1040900. eCollection 2022. Front Microbiol. 2022. PMID: 36466681 Free PMC article. - _S_-Adenosylmethionine: more than just a methyl donor.
Lee YH, Ren D, Jeon B, Liu HW. Lee YH, et al. Nat Prod Rep. 2023 Sep 20;40(9):1521-1549. doi: 10.1039/d2np00086e. Nat Prod Rep. 2023. PMID: 36891755 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- 21632007, 21502217 and 21621002/National Natural Science Foundation of China (National Science Foundation of China)/International
- XDB20000000, QYZDJ-SSW-SLH037/Chinese Academy of Sciences (CAS)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources