Experimentally induced metamorphosis in highly regenerative axolotl (ambystoma mexicanum) under constant diet restructures microbiota - PubMed (original) (raw)
Experimentally induced metamorphosis in highly regenerative axolotl (ambystoma mexicanum) under constant diet restructures microbiota
Turan Demircan et al. Sci Rep. 2018.
Abstract
Axolotl (Ambystoma mexicanum) is a critically endangered salamander species and a model organism for regenerative and developmental biology. Despite life-long neoteny in nature and in captive-bred colonies, metamorphosis of these animals can be experimentally induced by administering Thyroid hormones (THs). However, microbiological consequences of this experimental procedure, such as host microbiota response, remain largely unknown. Here, we systematically compared host bacterial microbiota associated with skin, stomach, gut tissues and fecal samples, between neotenic and metamorphic axolotls based on 16S rRNA gene sequences. Our results show that distinct bacterial communities inhabit individual organs of axolotl and undergo substantial restructuring through metamorphosis. Skin microbiota among others, shifted sharply, as highlighted by a major transition from Firmicutes-enriched to Proteobacteria-enriched relative abundance and precipitously decreased diversity. Fecal microbiota of neotenic and metamorphic axolotl shared relatively higher similarity, suggesting that diet continues to shape microbiota despite fundamental transformations in the host digestive organs. We also reproduced the previous finding on reduction in regenerative capacity in limbs of axolotl following metamorphosis, highlighting the need to investigate whether shifts in microbiota is causally linked to regenerative capacity of axolotl. The initial results on axolotl microbiota provide novel insights into microbiological aspects of axolotl metamorphosis and will establish a baseline for future in-depth studies.
Conflict of interest statement
The authors declare no competing interests.
Figures
Figure 1
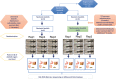
Experimental design for the comparison of neotenic and metamorphic axolotl. Of 48 siblings, a subset of 24 axolotls (9 axolotls for metamorphosis and 15 axolotls for regeneration experiments) were randomly selected and induced metamorphosis by T4 hormone administration while the rest kept untreated in neoteny. 30 animals (15 neotenic and 15 metamorphosed) were used in regeneration experiments and 18 animals (9 neotenic and 9 metamorphic) were housed individually for microbiome analysis. Each sample groups for skin, gut, stomach and fecal samples consisted of 3 replicates following randomization and pooling.
Figure 2
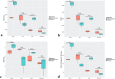
Effect of metamorphosis on bacterial diversity between neotenic and metamorphic axolotl. Box plots illustrate the comparison of diversity indices; Observed (a), Chao1 (b), Shannon (c) and Faith’s Phylogenetic Diversity (PD) measures.
Figure 3
Beta diversity analysis based on Bray-Curtis distance matrix showing separation of neotenic and metamorphic bacterial communities. Samples were compared using PCO (a) and Canonical Analysis of Principal Coordinates (CAP) (b) methods.
Figure 4
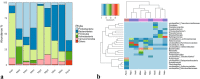
Mean relative abundances of 16S rRNA sequences. Phylum level relative abundance as bar chart (a), genus level relative abundances shown as heatmap (Individual taxa displayed if the its abundance in any sample ≥5%). Samples and bacterial taxa were clustered using average linkage hierarchical clustering of a distance matrix based on Bray–Curtis distance and taxa abundances, respectively. Samples from each group were color coded on the column side bar as follows: aqua samples (brown); samples from neotenic axolotl (light slate blue), metamorphic axolotl (magenta) (b).
Figure 5
Differentially enriched genus level taxa and indicator species in samples from neotenic and metamorphic axolotl. The color scale bar indicates log2 fold changes for absolute OTU abundances (DESeq2 analysis (q < 0.01) (**a**), Bubble plot showing indicator species. Only highly significant indicator values (IndVal >0.7, q < 0.01) are displayed. Size of bubble symbol is proportional to the mean relative abundance of indicator OTUs and the color scale bar shows indicator value for each OTU (b).
Figure 6
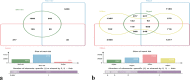
Venn diagrams showing the number of unique and shared OTUs. Skin and aqua samples (a), and gut and fecal samples (b), collected from neotenic and metamorphic axolotls as indicated in the diagram.
Similar articles
- A histological atlas of the tissues and organs of neotenic and metamorphosed axolotl.
Demircan T, İlhan AE, Aytürk N, Yıldırım B, Öztürk G, Keskin İ. Demircan T, et al. Acta Histochem. 2016 Sep;118(7):746-759. doi: 10.1016/j.acthis.2016.07.006. Epub 2016 Jul 18. Acta Histochem. 2016. PMID: 27436816 - Comparison of protein expression profile of limb regeneration between neotenic and metamorphic axolotl.
Sibai M, Altuntaş E, Süzek BE, Şahin B, Parlayan C, Öztürk G, Baykal AT, Demircan T. Sibai M, et al. Biochem Biophys Res Commun. 2020 Feb 5;522(2):428-434. doi: 10.1016/j.bbrc.2019.11.118. Epub 2019 Nov 22. Biochem Biophys Res Commun. 2020. PMID: 31767146 - Forever young: Endocrinology of paedomorphosis in the Mexican axolotl (Ambystoma mexicanum).
De Groef B, Grommen SVH, Darras VM. De Groef B, et al. Gen Comp Endocrinol. 2018 Sep 15;266:194-201. doi: 10.1016/j.ygcen.2018.05.016. Epub 2018 May 16. Gen Comp Endocrinol. 2018. PMID: 29777689 Review. - What mechanisms control neoteny and regulate induced metamorphosis in urodeles?
Rosenkilde P, Ussing AP. Rosenkilde P, et al. Int J Dev Biol. 1996 Aug;40(4):665-73. Int J Dev Biol. 1996. PMID: 8877439 Review.
Cited by
- Construction of the axolotl cell landscape using combinatorial hybridization sequencing at single-cell resolution.
Ye F, Zhang G, E W, Chen H, Yu C, Yang L, Fu Y, Li J, Fu S, Sun Z, Fei L, Guo Q, Wang J, Xiao Y, Wang X, Zhang P, Ma L, Ge D, Xu S, Caballero-Pérez J, Cruz-Ramírez A, Zhou Y, Chen M, Fei JF, Han X, Guo G. Ye F, et al. Nat Commun. 2022 Jul 22;13(1):4228. doi: 10.1038/s41467-022-31879-z. Nat Commun. 2022. PMID: 35869072 Free PMC article. - Proteome data to explore the axolotl limb regeneration capacity at neotenic and metamorphic stages.
Demircan T, Sibai M, Altuntaş E. Demircan T, et al. Data Brief. 2020 Jan 28;29:105179. doi: 10.1016/j.dib.2020.105179. eCollection 2020 Apr. Data Brief. 2020. PMID: 32055664 Free PMC article. - Ultrasound description of the coelomic cavity of the axolotl (Ambystoma mexicanum) in a clinically healthy population: a pilot study.
Vieu S, Le Poul N, Tur L, Aupée C, Kerbrat-Copy R, Bouhsina N, Cojean O, Fusellier M. Vieu S, et al. Sci Rep. 2024 May 23;14(1):11787. doi: 10.1038/s41598-024-62264-z. Sci Rep. 2024. PMID: 38782987 Free PMC article. - Altered metabolic state impedes limb regeneration in salamanders.
Peng ZL, Yin BX, Ren RM, Liao YL, Cai H, Wang H. Peng ZL, et al. Zool Res. 2021 Nov 18;42(6):772-782. doi: 10.24272/j.issn.2095-8137.2021.186. Zool Res. 2021. PMID: 34643071 Free PMC article. - Screening Salamanders for Symbionts.
Vickers E, Kerney R. Vickers E, et al. Methods Mol Biol. 2023;2562:425-442. doi: 10.1007/978-1-0716-2659-7_28. Methods Mol Biol. 2023. PMID: 36272092
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases