Programmed loading and rapid purification of engineered bacterial microcompartment shells - PubMed (original) (raw)
Programmed loading and rapid purification of engineered bacterial microcompartment shells
Andrew Hagen et al. Nat Commun. 2018.
Abstract
Bacterial microcompartments (BMCs) are selectively permeable proteinaceous organelles which encapsulate segments of metabolic pathways across bacterial phyla. They consist of an enzymatic core surrounded by a protein shell composed of multiple distinct proteins. Despite great potential in varied biotechnological applications, engineering efforts have been stymied by difficulties in their isolation and characterization and a dearth of robust methods for programming cores and shell permeability. We address these challenges by functionalizing shell proteins with affinity handles, enabling facile complementation-based affinity purification (CAP) and specific cargo docking sites for efficient encapsulation via covalent-linkage (EnCo). These shell functionalizations extend our knowledge of BMC architectural principles and enable the development of minimal shell systems of precisely defined structure and composition. The generalizability of CAP and EnCo will enable their application to functionally diverse microcompartment systems to facilitate both characterization of natural functions and the development of bespoke shells for selectively compartmentalizing proteins.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
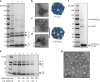
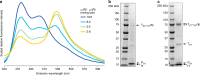
Comparison of different shell preparation methods. a SDS-PAGE analysis of HO shell preparations. Lane 1: Classic, Lane 2: in vivo capped, Lane 3: ex vivo capped. Shell protein identities indicated by arrows; loading normalized to A280 readings. b–d Negative stain TEM micrographs of three different shell preparations and corresponding structural models. Scale bar = 100 nm. e SDS-PAGE analysis of eluates from pentamer titration experiment. Lane 1: Ex vivo capped shells from Fig. 1a, Lanes 2–6: Titration of pentamers with equivalent volumes of each culture used in mixing experiments expressed in milliliters. f SDS-PAGE analysis of Halo carboxysome shell preparation using affinity purification. g Negative stain TEM micrographs of Halo carboxysome shell preparation. Scale bar = 50 nm. Results presented are representative of two independent biological replicates for a–d, f, and g. Findings in figure (e) recapitulate pilot experiments in which a subset of specified lysate ratios were used
Fig. 2
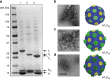
Comparison of different minimal shells. a SDS-PAGE analysis of crude shell preparations. Lane 1: HT1PSII Lane 2: HT2PSII Lane 3: HT3PSII. Shell protein identities indicated by arrows. b–d Negative stain TEM micrographs of three minimal shell preparations and corresponding structural models. Scale bar = 100 nm
Fig. 3
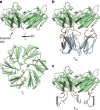
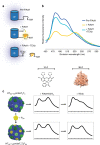
Molecular models of T1, TSC and TST subunits. a Model of T1 (wt) (PDB: 5DIH) as viewed from side of shell (top) and lumen (bottom). b and c Models of TSC and TST (respectively) viewed from the side. Flexible coil regions colored in brown; SpyCatcher and SpyTag regions colored in gray
Fig. 4
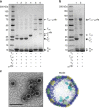
SDS-PAGE and electron micrographs of various shell preparations. a, b SDS-PAGE of shell preparations. Composition of shell preparations given in tabular form below each lane. c Negative stain TEM of HTSC~STcfpPSII shell preparation (lane 6 in figure a) and cutaway model of shells (cfp rendered in turquoise, not completely functionalized for clarity). Results presented are representative of at least two independent biological replicates of each sample preparation
Fig. 5
Fluorescence spectra and SDS-PAGE analysis of shells containing ex vivo programmed cargo. a Scaled emission spectra (excitation: 405 nm) of programmed cargo. STyfp-only trace (0:10) was not plotted—in the absence of a FRET donor, the fluorescence signal is negligible. b SDS-PAGE analysis of STcfp (10:0) programmed shells. c SDS-PAGE analysis of SCcfp programmed shells. Results are representative of two independent biological replicates
Fig. 6
Probe properties and permeability assay of uncapped and capped shells. a Schematic of probe behavior in presence of FlAsH and TEV protease. b Fluorescence emission spectra (excitation: 405 nm) of unencapsulated probe. c Emission spectra (450–600 nm) of encapsulated probe in uncapped and capped shells, in the presence of FlAsH and TEV protease. Axis numbers omitted for clarity; tick marks correspond to numbers in b. Results are representative of two independent technical replicates. Bacterial microcompartments are protein-bound organelles encapsulating segments of metabolic pathways. Here the authors functionalize shell proteins to facilitate facile purification and enable cargo encapsulation via covalent linkage
Similar articles
- Heterologous Assembly of Pleomorphic Bacterial Microcompartment Shell Architectures Spanning the Nano- to Microscale.
Ferlez BH, Kirst H, Greber BJ, Nogales E, Sutter M, Kerfeld CA. Ferlez BH, et al. Adv Mater. 2023 Jun;35(23):e2212065. doi: 10.1002/adma.202212065. Epub 2023 Apr 25. Adv Mater. 2023. PMID: 36932732 Free PMC article. - Modulation of Hybrid GRM2-type Bacterial Microcompartment Shells through BMC-H Shell Protein Fusion and Incorporation of Non-native BMC-T Shell Proteins.
Česle EEL, Ta Rs K, Jansons J, Kalniņš G. Česle EEL, et al. ACS Synth Biol. 2023 Nov 17;12(11):3275-3286. doi: 10.1021/acssynbio.3c00281. Epub 2023 Nov 8. ACS Synth Biol. 2023. PMID: 37937366 - Structural Characterization of a Synthetic Tandem-Domain Bacterial Microcompartment Shell Protein Capable of Forming Icosahedral Shell Assemblies.
Sutter M, McGuire S, Ferlez B, Kerfeld CA. Sutter M, et al. ACS Synth Biol. 2019 Apr 19;8(4):668-674. doi: 10.1021/acssynbio.9b00011. Epub 2019 Mar 27. ACS Synth Biol. 2019. PMID: 30901520 Free PMC article. - The shells of BMC-type microcompartment organelles in bacteria.
Yeates TO, Jorda J, Bobik TA. Yeates TO, et al. J Mol Microbiol Biotechnol. 2013;23(4-5):290-9. doi: 10.1159/000351347. Epub 2013 Aug 5. J Mol Microbiol Biotechnol. 2013. PMID: 23920492 Free PMC article. Review. - Biotechnological Advances in Bacterial Microcompartment Technology.
Lee MJ, Palmer DJ, Warren MJ. Lee MJ, et al. Trends Biotechnol. 2019 Mar;37(3):325-336. doi: 10.1016/j.tibtech.2018.08.006. Epub 2018 Sep 17. Trends Biotechnol. 2019. PMID: 30236905 Review.
Cited by
- Characterization of a widespread sugar phosphate-processing bacterial microcompartment.
Dwyer ME, Sutter M, Kerfeld CA. Dwyer ME, et al. Commun Biol. 2024 Nov 24;7(1):1562. doi: 10.1038/s42003-024-07287-y. Commun Biol. 2024. PMID: 39580597 Free PMC article. - A robust synthetic biology toolkit to advance carboxysome study and redesign.
Trettel DS, Hoang Y, Vecchiarelli AG, Gonzalez-Esquer CR. Trettel DS, et al. bioRxiv [Preprint]. 2024 Oct 8:2024.10.08.617227. doi: 10.1101/2024.10.08.617227. bioRxiv. 2024. PMID: 39416180 Free PMC article. Preprint. - Bacterial microcompartments as a next-generation metabolic engineering tool: utilizing nature's solution for confining challenging catabolic pathways.
Doron L, Kerfeld CA. Doron L, et al. Biochem Soc Trans. 2024 Jun 26;52(3):997-1010. doi: 10.1042/BST20230229. Biochem Soc Trans. 2024. PMID: 38813858 Free PMC article. Review. - Overcoming Symmetry Mismatch in Vaccine Nanoassembly through Spontaneous Amidation.
Rahikainen R, Rijal P, Tan TK, Wu HJ, Andersson AC, Barrett JR, Bowden TA, Draper SJ, Townsend AR, Howarth M. Rahikainen R, et al. Angew Chem Weinheim Bergstr Ger. 2021 Jan 4;133(1):325-334. doi: 10.1002/ange.202009663. Epub 2020 Oct 26. Angew Chem Weinheim Bergstr Ger. 2021. PMID: 38504824 Free PMC article. - Modeling bacterial microcompartment architectures for enhanced cyanobacterial carbon fixation.
Trettel DS, Pacheco SL, Laskie AK, Gonzalez-Esquer CR. Trettel DS, et al. Front Plant Sci. 2024 Feb 15;15:1346759. doi: 10.3389/fpls.2024.1346759. eCollection 2024. Front Plant Sci. 2024. PMID: 38425792 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI114975/AI/NIAID NIH HHS/United States
- 1R01AI114975-01/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)/International
- FG02-91ER20021/DOE | SC | Basic Energy Sciences (BES)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous