The Role of m6A/m-RNA Methylation in Stress Response Regulation - PubMed (original) (raw)
. 2018 Jul 25;99(2):389-403.e9.
doi: 10.1016/j.neuron.2018.07.009.
Carola Eggert 1, Paul M Kaplick 1, Matthias Eder 1, Simone Röh 2, Lisa Tietze 1, Christian Namendorf 3, Janine Arloth 2, Peter Weber 2, Monika Rex-Haffner 2, Shay Geula 4, Mira Jakovcevski 5, Jacob H Hanna 4, Dena Leshkowitz 6, Manfred Uhr 3, Carsten T Wotjak 1, Mathias V Schmidt 1, Jan M Deussing 1, Elisabeth B Binder 7, Alon Chen 8
Affiliations
- PMID: 30048615
- PMCID: PMC6069762
- DOI: 10.1016/j.neuron.2018.07.009
The Role of m6A/m-RNA Methylation in Stress Response Regulation
Mareen Engel et al. Neuron. 2018.
Abstract
N6-methyladenosine (m6A) and N6,2'-O-dimethyladenosine (m6Am) are abundant mRNA modifications that regulate transcript processing and translation. The role of both, here termed m6A/m, in the stress response in the adult brain in vivo is currently unknown. Here, we provide a detailed analysis of the stress epitranscriptome using m6A/m-seq, global and gene-specific m6A/m measurements. We show that stress exposure and glucocorticoids region and time specifically alter m6A/m and its regulatory network. We demonstrate that deletion of the methyltransferase Mettl3 or the demethylase Fto in adult neurons alters the m6A/m epitranscriptome, increases fear memory, and changes the transcriptome response to fear and synaptic plasticity. Moreover, we report that regulation of m6A/m is impaired in major depressive disorder patients following glucocorticoid stimulation. Our findings indicate that brain m6A/m represents a novel layer of complexity in gene expression regulation after stress and that dysregulation of the m6A/m response may contribute to the pathophysiology of stress-related psychiatric disorders.
Keywords: Fto; Mettl3; RNA modification; m(6)A; m(6)Am; major depressive disorder; post-transcriptional regulation; stress.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
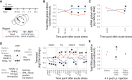
Mapping the Transcriptome-wide m6A/m Landscape after Acute Restraint Stress in the Mouse Cortex Using m6A/m-Seq FC, fold change; TE, translation efficiency. (A) Approximately half of the genes expressed in the mouse cortex are m6A/m methylated, but only a minor fraction is regulated by acute stress on cortex-wide level. m6A/m-seq of mouse cortex poly(A)-RNA basal or 4 hr after 15 min restraint stress; n = 6–7, each pooled from 3 mice. Stress-regulated m6A/m peaks, Q < 0.1 and absolute log2 fold change > 0.2; stress-regulated mRNAs ( = differential expressed genes), Q < 0.1 and absolute log2 fold change > 0.1. (B) m6A/m peaks are enriched at the 5′ UTR and the stop codon with similar distribution of all and stress-regulated peaks (peak distribution mapped along mRNA relative position). (C) GGACWB is the most abundant motif detected in m6A/m peaks and enriched at peak summits. Top enriched sequence motif and its position across the detected m6A/m peaks. GGAC was detected in 84%, GGACWB in 63% of the peaks. (D) Two examples of stress-regulated m6A/m peaks and replication of their quantitative regulation by m6A/m-RNA immunoprecipitation (RIP)-qPCR in an unrelated cohort of animals. Left panel per gene: averaged sequence tracks and peaks; arrows indicate quantitatively regulated peaks. Right panel per gene: differential methylation was validated in a separate cohort of mice using full-length m6A/m-RIP-qPCR, including an intermediated time point (1 hr). n = 7, mean ± SEM; asterisks [∗] depict omnibus Tukey post hoc tests to basal p < 0.05 after FDR-corrected one-way ANOVA. (E) Bioinformatic dissection of m6Am peaks based on their position at the transcription start site, observing 1,801 putative m6Am sites. m6Am peaks do not show a preference for stress-regulated peaks. (F) Regulation of translation efficiency (TE assessed by ribosome profiling) by stress does not correlate well or overlap with regulation of m6A/m methylation. n = 6 for ribosome profiling; n = 6–7 for m6A/m profiling. Shown are fold changes upon stress using only genes abundantly detected in ribosome profiling sequencing with significance determined by Q < 0.1 and absolute log2 fold change > 0.2. See also Figures S1 and S2 and Table S1.
Figure 2
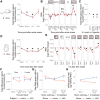
Acute Restraint Stress Regulates Brain Global m6A/m and Expression of the m6A/m Regulatory System in a Time- and Region-Specific Manner (A) Experimental design. PFC, medial prefrontal cortex (orange); AMY, central and basolateral amygdala (blue). (B) Global m6A/m is decreased in the PFC and increased in the AMY after acute restraint stress. Global m6A/m assay on total RNA, n = 12, mean ± SEM; two-way ANOVA interaction effect F(4, 110) = 24.045, p < 0.001. Asterisk (∗) depicts omnibus Tukey post hoc tests to basal p < 0.05. Results were replicated in three independent mouse cohorts with only one experiment shown. (C) Likewise, global mRNA m6A is decreased when measured with LC-MS/MS. n = 7, mean ± SEM. Specific measurement of only m6A. Two-way ANOVA interaction effect F(1, 24) = 159.537, p < 0.001. Asterisk (∗) depicts omnibus Tukey post hoc tests to basal p < 0.05. (D) m6A/m regulatory genes _Mettl3_, _Fto_, _Alkbh5_, and _Ythdc1_ are differentially expressed after acute stress in the brain. See also Figure S3. n = 12, log2 fold change ± SEM; two-way MANOVA, significant interaction effects for _Fto_, _Alkbh5_, and _Ythdc1_; main stress effect for _Mettl3_; each FDR-corrected p < 0.05 and n2 > 0.01. Asterisk (∗) depicts omnibus Tukey post hoc tests to basal p < 0.05. See also Table S2. Results were replicated in three independent mouse cohorts with only one experiment shown. (E) Global m6A/m is decreased in the PFC and increased in the AMY after corticosterone i.p. injection, but not after dexamethasone injection. Corticosterone, 250 μg/ kg; dexamethasone, 10 mg/kg. Global m6A/m assay on total RNA, n = 12, mean ± SEM. Two-way ANOVA reported a significant interaction effect (F(4, 96) = 12.887, p < 0.001). Asterisk (∗) indicates omnibus Tukey post hoc tests p < 0.05 compared to area basal. See also Figure S3.
Figure 3
Absolute Regulation of m6A/m Methylation Is Site Specific (A) A synthetic RNA oligonucleotide with three internal m6A/m sites was used for validation and internal normalization of the m6A/m-RIP-qPCR. See also Figure S4. (B) m6A/m-RIP-qPCR detects the methylated spike-in oligonucleotide in a linear fashion without impairing precipitation efficiency for endogenous transcripts in the concentration range used for experiments. Methylated spike-in oligo was added to unfragmented total RNA and precipitated with anti-m6A/m antibody (m6A/m-RIP) or rabbit IgG (IgG NC). n = 3 technical replicates, normalized expression to 1 fmol input control. Mean ± SEM. (C) m6A/m-RIP-qPCR accurately quantifies differential methylation of the spike-in oligo. Spike-in oligo (1 fmol) mixed from fully methylated and fully unmethylated spike-in was added to unfragmented total RNA and precipitated with m6A/m-RIP-qPCR. n = 3 technical replicates, normalized to input control. Mean ± SEM. (D) Absolute full-length m6A/m levels of stress-related and synaptic plasticity-related transcripts are differentially regulated in the PFC and AMY of stress-related candidate transcripts and synaptic-plasticity-related candidate transcripts after stress. See also Figure S4. n = 8, mean ± SEM. Significant effects observed in FDR-corrected two-way MANOVA (p < 0.05, n2 > 0.01) are coded in the rows “m6A/m stress effect” and “RNA stress effect.” Orange/blue arrows, PFC-/AMY-specific stress effect (interaction effect two-way ANOVA, one-way follow-up significant in respective tissue); black arrow, stress main effect; equals sign, no interaction or stress main effect in two-way ANOVA. See also Table S2. (E) The majority of transcripts measured are expressed or regulated in a region-specific manner. Percent of transcripts with significant interaction or main effect in FDR-corrected 2×2 MANOVA. (F) Stress regulation of m6A/m negatively correlates with changes in RNA levels. log2 fold changes of m6A/m and RNA after stress to basal time points, n = 44 per group; black line, linear model + 95% CI. For generalized linear models (GLMs), see Table S2. (G) General patterns of m6A/m changes vary in extent and direction depending on brain region and time point. Density plots of data depicted in (D); t test. (H) The m6A/m change at the 1 hr time point correlates with the m6A/m change at 4 hr in the PFC, but not AMY, indicating that in the PFC, m6A/m change 1 hr after stress is a proxy for later change. Orange line, linear model for PFC only + 95% CI. For GLMs, see Table S2. See also Figure S4.
Figure 4
Depletion of METTL3 and FTO in Adult Excitatory Neurons Using the Camk2a-Cre Driver Changes the Cortex Epitranscriptome (A) Mettl3 and Fto mRNA are depleted from the neocortex and hippocampus in Mettl3 cKO and Fto cKO mice, respectively. In situ hybridization, n = 3, representative shown. WT, wild-type; cKO, conditional. (B) METTL3 and FTO proteins are significantly depleted in Mettl3 cKO and Fto cKO mice, respectively. Western blot of PFC protein, optical density normalized from digitally acquired image signal normalized to ACTB protein. n = 4–5, mean ± SEM. ∗p < 0.05, t test. For full blots, see Figure S5. (C) Global mRNA m6A is decreased in Mettl3 cKO mice, but not in Fto cKO mice, when measured with LC-MS/MS. n = 5, mean ± SEM, m6A-specific measurement. Two-way ANOVA interaction effect F(1, 19) = 106.269, p < 0.001. ∗p < 0.05, omnibus Tukey post hoc tests to respective WT. See also Figure S5. (D) Global mRNA m6Am is increased in Fto cKO mice when measured with LC-MS/MS. n = 5, mean ± SEM, m6Am-specific measurement. Data are shown relative to Am, which is not altered in Fto cKO mice. ∗p < 0.05, t test. For LC-MS/MS traces, see Figure S5. (E) The m6A/m epitranscriptome is widely altered in in Mettl3 cKO and Fto cKO mice. m6A/m-seq on mouse cortex poly(A)-RNA of WT and cKO animals reported 1,266 and 78 significantly different methylated m6A/m peaks in Mettl3 cKO and in Fto cKO compared to WT, respectively, with 14 shared peaks. n = 3–5, each pooled from 3 mice. WT of both lines were grouped together as we observed no major regulation between them. Shown are log2 fold changes of methylation in cKO relative to WT mice using 10,109 high-confidence consensus m6A/m peaks detected across all groups, mapping to 6,056 unique genes. Significantly regulated m6A/m peaks are Q < 0.1 and absolute log2 fold change > 0.5. (F) m6A/m peaks are enriched at the stop codon with a less prominent enrichment at the 5′ UTR, as observed in Figure 1. Differentially methylated peaks in both Mettl3 cKO and Fto cKO mice show an increased preference for 5′ UTR position with a decreased preference for CDS peaks in Mettl3 cKO differential peaks. Peak distribution mapped along mRNA relative position. (G) Two examples m6A/m peaks regulated only in Mettl3 cKO or in both Mettl3 cKO and Fto cKO. Shown are averaged sequence tracks m6A/m-seq and RNA-seq per group and detected m6A/m peaks. Arrows indicate quantitatively regulated peaks (Q < 0.1, absolute log2 fold change >0.5). See also Figure S5 and Table S3.
Figure 5
Deletion of Mettl3 or Fto in Adult Excitatory Neurons of the Hippocampus CA1 and CA3 via a Nex-CreERT2 Driver Line and Knockout Induction in Adult Animals via Tamoxifen Administration Alters Gene Expression in Animals (A) Mettl3 and Fto mRNA are depleted from the dorsal (d) and ventral (v) hippocampus CA1 and CA3 in Mettl3 cKO (blue) and Fto cKO (pink) mice, respectively. WT, wild-type; cKO, conditional knockout; DG, dentate gyrus. In situ hybridization; expression was quantified from digitalized films in arbitrary units (AU); mean ± SEM, n = 4 for Mettl3 WT and cKO, n = 11–14 for Fto WT and cKO, signal averaged across both hemispheres; ∗p < 0.05, t test. (B) METTL3 and FTO proteins are significantly depleted in Mettl3 cKO and Fto cKO mice, respectively. Protein was isolated from dissected dCA1/dCA3 and measured by western blot normalized to ACTB protein. n = 3–4, optical density normalized from digitally acquired images, mean ± SEM. ∗p < 0.05, t test. For full blots, see Figure S6. (C) mRNA-seq of adult CA1 and CA3 shows altered gene expression after deletion of _Mettl3_ and _Fto_ in non-stressed basal animals. More genes are differentially expressed after deletion of _Mettl3_ (84) compared to deletion of _Fto_ (15), with very few overlapping (3). log2 change by DESeq2 baseMean gene abundance from RNA-seq of adult basal animals. Differentially expressed by colored dots and in Venn circles, Q < 0.1, log2 fold change > 0.5. (D) Four representative examples of genes expressed in a knockout × genotype-specific pattern. In total, 104 genes were found to be expressed in a knockout × genotype interaction-dependent matter. Normalized counts relative to Mettl3 WT. n = 5. See also Figures S6 and S7 and Table S4.
Figure 6
Animals with Adult Excitatory Neuron-Specific Depletion of Mettl3 and Fto Using a Nex-CreERT2 Driver Line Have Impaired Fear Coping, Differential Transcriptomic Response to Fear, and Changes in Hippocampus CA1 Electrophysiological Properties (A) Both Mettl3 cKO and Fto cKO animals display increased conditioned fear memory long-term maintained during fear extinction. The primary fear response was not altered. Fto cKO animals also have increased contextual fear memory. No difference was observed in the Y-maze test or the object recognition test (ORT). CS, conditioned stimulus; lightning bolt, US, unconditioned stimulus; Ext, extinction. n = 11–13, mean ± SEM. Fear expression was binned in 1 min intervals during CS representation. Asterisk (∗) depicts a main genotype effect in repeated-measurements ANOVA for CS and Ext bins and a t test p < 0.05 for all other data points. (B) The transcriptomic response 24 hr after fear conditioning (FC) is altered in both animals with _Mettl3_ or _Fto_ depletion. log2 RNA fold change in WT versus cKO animals of only those genes with a significant genotype × FC effect. Q < 0.1, absolute log2 fold change > 0.5, n = 5. (C) More genes express a genotype-dependent FC effect in Fto cKOs compared to Mettl3 cKOs with low overlap. Four examples of such genes are shown. Significant genotype × FC in the examples is depicted by blue (Mettl3 cKOs) and pink (Fto cKOs) opposite arrows. Q < 0.1, absolute log2 fold change > 0.5, n = 5. (D) Long-term potentiation (LTP), but not short-term plasticity, in CA1 was attenuated in Fto cKO mice, but not Mettl3 cKO mice. Short-term synaptic plasticity was measured by paired-pulse facilitation (PPF). n = 10–12 slices from 5–6 animals, mean ± SEM plus representative LTP trace curves; HFS, high-frequency stimulation. ∗p < 0.05, t test, on the average field excitatory postsynaptic potential (fEPSP) slope 50–60 min post-HFS. See also Figures S6 and S7 and Table S4.
Figure 7
Global m6A/m in Blood Is Transiently Decreased after Stress in Mice and after Stimulation with GCs in Healthy Humans, but This Glucocorticoid-Induced m6A/m Reduction Is Absent in Blood and BLCLs from MDD Patients (A) Global m6A/m is transiently decreased in mouse blood after acute stress. Global m6A/m assay on total RNA, n = 8, mean ± SEM. Asterisks (∗) depict omnibus post hoc comparisons to basal, p < 0.05, after Kruskal-Wallis test, p < 0.05. (B) Global m6A/m changes in mouse blood are accompanied by changes in m6A/m regulatory genes. qPCR on total mouse blood, log2 fold changes of different genes to basal. n = 8, mean ± SEM. Red colored gene names, one-way ANOVA. Asterisks (∗) depict omnibus Tukey post hoc tests to basal p < 0.05; see Table S2. (C) Furthermore, global m6A/m is decreased in mouse blood after both corticosterone and dexamethasone i.p. injection. Corticosterone, 250 μg/kg; dexamethasone, 10 mg/kg. Global m6A/m assay on total RNA, n = 12, mean ± SEM. Two-way ANOVA reported a significant interaction effect (F(4, 96) = 12.887, p < 0.001). Stars indicate omnibus Tukey post hoc tests, p < 0.05 compared to area basal. (D) In a similar way, global m6A/m is temporarily decreased in the blood of healthy human subjects after treatment with 1.5 mg dexamethasone (Dex). Global m6A/m assay on total whole blood RNA, n = 25 healthy men, mean ± SEM. Kruskal-Wallis test, p < 0.001. Asterisks (∗) depict omnibus Tukey post hoc tests to basal p < 0.05. (E) Expression of m6A/m regulatory genes in human blood is also affected by dexamethasone. Microarray of human whole blood at baseline and 3 hr after intake of Dex, n = 160 mixed healthy and diseased subjects, mean ± SEM. Asterisks (∗) depict Bonferroni-corrected t tests to basal p < 0.05. (F) The dexamethasone-induced m6A/m decrease in human blood m6A/m is absent in MDD patients. n = 25, male and female, healthy and MDD subjects each, mean ± SEM. Three-way mixed-design ANOVA, significant interaction effect of treatment and subject status (F(1, 96) = 11.184, p = 0.001), but no interaction with sex. Asterisks (∗) depict omnibus Tukey post hoc tests to sex basal p < 0.05. (G) Global m6A/m is decreased in B lymphocyte cell lines (BLCLs) in a concentration-dependent manner after 1 hr treatment with cortisol. Global m6A/m assay on total RNA, n = 5 biological replicates with 3 technical replicates each, mean ± SEM. Two-way ANOVA, significant interaction effect of cortisol and donor status (F(3, 24) = 44.365, p < 0.001). Asterisks (∗) depict omnibus Tukey post hoc tests to basal p < 0.05. (H) The same regulation is observed on mRNA m6A using LC-MS/MS. n = 5, mean ± SEM. Specific detection of m6A. Two-way ANOVA, significant interaction effect of cortisol and donor status (F(1, 20) = 19.196, p < 0.001). Asterisks (∗) depict omnibus Tukey post hoc tests to mock treatment p < 0.05. See also Figure S8.
Figure 8
m6A-Seq and m6A-RIP qPCR Reveal Donor-Specific Patterns of m6A/m Regulation Altered in Cells Obtained from Healthy and MDD Donors (A–E) The cortisol-responsive m6A/m epitranscriptome of healthy and MDD donor BLCLs was analyzed using m6A/m-seq comparing 1 hr mock-treated or 100 nM cortisol (Cort) conditions. We found 17,655 consensus high-confidence m6A/m peaks across all samples mapping to 8,681 genes. m6A/m-seq of each n = 3 BLCLs from healthy and MDD donors, each after 1 hr of mock or cortisol treatment. (A) m6A/m peaks in BLCLs are enriched in genes with stress-responsive and metabolic functions. Fifteen highest enriched biological process GO terms with FDR-corrected Q < 0.1. (B) Differential m6A/m and gene expression analysis in BLCLs reveals significant regulation of methylation after cortisol treatment with many m6A/m peaks regulated by cortisol in a donor-specific fashion and almost absent effects by donor status alone. Gene expression was less regulated than m6A/m regulation, lacking any donor-specific effects by cortisol. Number of significantly regulated peaks/genes with Q < 0.1 and absolute log2 fold change > 0.5, reporting 2 × 2 and post hoc effects within the single donor groups. Peaks and genes with significant interaction effects are removed from main treatment and main donor effect. (C) Many peaks are regulated by cortisol in a donor-specific fashion. log2 fold changes of m6A/m peaks by cortisol within each donor group; significant peaks defined as above. (D) m6A/m peaks in BLCLs show similar peak position enrichments as m6A/m peaks in mouse brain with preference for cortisol-regulated peaks at 5′ UTR and CDS. Peak distribution mapped along mRNA relative position. (E) m6A/m peaks regulated by cortisol alone (main effect) have higher fold changes in healthy donors compared to BLCLs from MDD donors. (F) Assessing absolute transcript methylation using m6A/m-RIP-qPCR, the cortisol-responsive genes FKBP5, IRS2, and TSC22D3 m6A/m were found specifically downregulated in cell lines of healthy, but not MDD, donors after stimulation with cortisol (cort). m6A/m-RIP-qPCR. n = 5, mean ± SEM. Significant effects observed in FDR-corrected two-way MANOVA (p < 0.05) are coded in the rows “m6A/m Cort effect” and “RNA Cort effect.” Orange arrows, healthy donor-specific Cort effect (interaction effect two-way ANOVA, one-way follow-up significant in healthy donors only); black arrow, Cort main stress effect; equals sign, no interaction or stress main effect in two-way ANOVA. For full statistics, see Table S2. (G) Density plots of m6A/m change upon cortisol treatment. Density plots of log2 fold change data as m6A/m-RIP-qPCR data depicted in (F); donor-dependent distributions of fold changes were compared using a t test. See also Figure S9 and Table S5.
Comment in
- Epitranscriptomes in the Adult Mammalian Brain: Dynamic Changes Regulate Behavior.
Yoon KJ, Ming GL, Song H. Yoon KJ, et al. Neuron. 2018 Jul 25;99(2):243-245. doi: 10.1016/j.neuron.2018.07.019. Neuron. 2018. PMID: 30048610 Free PMC article.
Similar articles
- Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice.
Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono V, Bredy TW. Widagdo J, et al. J Neurosci. 2016 Jun 22;36(25):6771-7. doi: 10.1523/JNEUROSCI.4053-15.2016. J Neurosci. 2016. PMID: 27335407 Free PMC article. - The emerging role of mRNA methylation in normal and pathological behavior.
Engel M, Chen A. Engel M, et al. Genes Brain Behav. 2018 Mar;17(3):e12428. doi: 10.1111/gbb.12428. Epub 2017 Nov 17. Genes Brain Behav. 2018. PMID: 29027751 Review. - m6 A RNA methylation: from mechanisms to therapeutic potential.
He PC, He C. He PC, et al. EMBO J. 2021 Feb 1;40(3):e105977. doi: 10.15252/embj.2020105977. Epub 2021 Jan 20. EMBO J. 2021. PMID: 33470439 Free PMC article. Review. - m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome.
Sun H, Li K, Zhang X, Liu J, Zhang M, Meng H, Yi C. Sun H, et al. Nat Commun. 2021 Aug 6;12(1):4778. doi: 10.1038/s41467-021-25105-5. Nat Commun. 2021. PMID: 34362929 Free PMC article. - _N_6-methyladenosine is required for the hypoxic stabilization of specific mRNAs.
Fry NJ, Law BA, Ilkayeva OR, Holley CL, Mansfield KD. Fry NJ, et al. RNA. 2017 Sep;23(9):1444-1455. doi: 10.1261/rna.061044.117. Epub 2017 Jun 13. RNA. 2017. PMID: 28611253 Free PMC article.
Cited by
- The RNA m5C methyltransferase NSUN1 modulates human malaria gene expression during intraerythrocytic development.
Tang R, Fan Y, Lu B, Jiang Q, Cheng X, Zhang Z, Shen L, Shang X. Tang R, et al. Front Cell Infect Microbiol. 2024 Oct 7;14:1474229. doi: 10.3389/fcimb.2024.1474229. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39435184 Free PMC article. - Global DNA and RNA Methylation Signature in Response to Antipsychotic Treatment in First-Episode Schizophrenia Patients.
Angelin M, Gopinath P, Raghavan V, Thara R, Ahmad F, Munirajan AK, Sudesh R. Angelin M, et al. Neuropsychiatr Dis Treat. 2024 Jul 19;20:1435-1444. doi: 10.2147/NDT.S466502. eCollection 2024. Neuropsychiatr Dis Treat. 2024. PMID: 39049939 Free PMC article. - Alternative cleavage and polyadenylation generates downstream uncapped RNA isoforms with translation potential.
Malka Y, Alkan F, Ju S, Körner PR, Pataskar A, Shulman E, Loayza-Puch F, Champagne J, Wenzel C, Faller WJ, Elkon R, Lee C, Agami R. Malka Y, et al. Mol Cell. 2022 Oct 20;82(20):3840-3855.e8. doi: 10.1016/j.molcel.2022.09.036. Mol Cell. 2022. PMID: 36270248 Free PMC article. - m6A modification of lncRNA in middle ear cholesteatoma.
He J, Xie S, Jin L, Fu J, Yuan Q, Liu W. He J, et al. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024 May 28;49(5):667-678. doi: 10.11817/j.issn.1672-7347.2024.230477. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 39174880 Free PMC article. - Altered Expression of the m6A Methyltransferase METTL3 in Alzheimer's Disease.
Huang H, Camats-Perna J, Medeiros R, Anggono V, Widagdo J. Huang H, et al. eNeuro. 2020 Sep 8;7(5):ENEURO.0125-20.2020. doi: 10.1523/ENEURO.0125-20.2020. Print 2020 Sep/Oct. eNeuro. 2020. PMID: 32847866 Free PMC article.
References
- Agarwal A., Dibaj P., Kassmann C.M., Goebbels S., Nave K.-A., Schwab M.H. In vivo imaging and noninvasive ablation of pyramidal neurons in adult NEX-CreERT2 mice. Cereb. Cortex. 2012;22:1473–1486. - PubMed
- Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous