Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis - PubMed (original) (raw)
. 2018 Sep 3;37(17):e99238.
doi: 10.15252/embj.201899238. Epub 2018 Jul 26.
Giovanni Quarato 3, Catherine Cloix 1 2, Jonathan Lopez 1 2, Jim O'Prey 1 2, Matthew Pearson 4, James Chapman 5, Hiromi Sesaki 6, Leo M Carlin 1 2, João F Passos 5 7, Ann P Wheeler 4, Andrew Oberst 8 9, Kevin M Ryan 1 2, Stephen Wg Tait 10 2
Affiliations
- PMID: 30049712
- PMCID: PMC6120664
- DOI: 10.15252/embj.201899238
Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis
Joel S Riley et al. EMBO J. 2018.
Abstract
During apoptosis, pro-apoptotic BAX and BAK are activated, causing mitochondrial outer membrane permeabilisation (MOMP), caspase activation and cell death. However, even in the absence of caspase activity, cells usually die following MOMP Such caspase-independent cell death is accompanied by inflammation that requires mitochondrial DNA (mtDNA) activation of cGAS-STING signalling. Because the mitochondrial inner membrane is thought to remain intact during apoptosis, we sought to address how matrix mtDNA could activate the cytosolic cGAS-STING signalling pathway. Using super-resolution imaging, we show that mtDNA is efficiently released from mitochondria following MOMP In a temporal manner, we find that following MOMP, BAX/BAK-mediated mitochondrial outer membrane pores gradually widen. This allows extrusion of the mitochondrial inner membrane into the cytosol whereupon it permeablises allowing mtDNA release. Our data demonstrate that mitochondrial inner membrane permeabilisation (MIMP) can occur during cell death following BAX/BAK-dependent MOMP Importantly, by enabling the cytosolic release of mtDNA, inner membrane permeabilisation underpins the immunogenic effects of caspase-independent cell death.
Keywords: BAX/BAK; apoptosis; cGAS‐STING; mitochondria; mtDNA.
© 2018 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
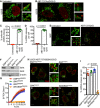
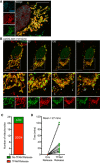
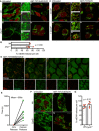
Figure 1. mtDNA is released from mitochondria following MOMP in a BAX/BAK‐dependent manner
- Fixed super‐resolution Airyscan images of U2OS cells immunostained with anti‐TOM20 (green) and anti‐DNA (red) antibodies. Scale bar = 10 μm. Representative images from three independent experiments.
- Airyscan images of U2OS cells treated with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh for 3 h, immunostained with anti‐TOM20 and anti‐DNA antibodies. Scale bar = 10 μm. Representative images from three independent experiments.
- Quantification of cells exhibiting > 10% mtDNA release following treatment with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh. Data are expressed as mean ± SD from three independent experiments and analysed by Student's _t_‐test.
- Quantification of the extent of mitochondrial DNA (mtDNA) nucleoid release per cell following treatment with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh. Data are expressed as mean ± SD from two independent experiments and analysed by Student's _t_‐test.
- Airyscan images of MEF cells untreated or treated with 10 μM ABT‐737 and 20 μM qVD‐OPh for 3 h, immunostained with anti‐TOM20 and anti‐DNA antibodies. Scale bar = 10 μm. Representative images from three independent experiments.
- BAX and BAK expression levels in U2OS cells with CRISPR‐Cas9‐mediated deletion of BAX, BAK or BAX/BAK.
- U2OS cells with BAX, BAK or BAX/BAK deletion by CRISPR‐Cas9 treated with 10 μM ABT‐737 and 2 μM S63845 and analysed for cell viability using an IncuCyte live‐cell imager and SYTOX Green exclusion. Data are expressed as mean ± SEM, representative of three independent experiments, and have been normalised to starting confluency.
- Airyscan images of U2OS (i) control cells or with CRISPR‐Cas9‐mediated deletion of either (ii) BAX, (iii) BAK or (iv) BAX and BAK treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh for 3 h. Scale bar = 10 μm. Representative images from three independent experiments.
- Quantification of mtDNA nucleoid release per cell in U2OS EMPTYCRISPR, BAX CRISPR, BAK CRISPR and BAX/BAK CRISPR cells. Data are expressed as mean ± SD from three independent experiments and analysed using Student's _t_‐test.
Source data are available online for this figure.
Figure EV1. BAX/BAK‐dependent MOMP commits cells to die
- U2OS cells treated with 10 μM ABT‐737 and 1 μM ActD ± 20 μM qVD‐OPh. Cell viability was analysed using an IncuCyte live‐cell imager and SYTOX Green exclusion. Data are expressed as mean ± SEM, representative of three independent experiments.
- Clonogenic survival assay of U2OS cells treated with 10 μM ABT‐737 and 2 μM S63845 ± 20 μM qVD‐OPh or 10 μM ABT‐737 and 1 μM ActD ± 20 μM qVD‐OPh. Representative images from three independent experiments.
- Airyscan images of U2OS cells treated with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh for 3 h, immunostained with anti‐TOM20 (green) and anti‐cytochrome c (red). Scale bar = 10 μm. Representative images from three independent experiments.
- Quantification of cytochrome c release from mitochondria. Data are expressed as mean ± SD from three independent experiments and analysed using Student's _t_‐test.
- Quantification of cytochrome c release from BAX‐, BAK‐, and BAX/BAK‐deleted cells. Data are expressed as mean ± SD from three independent experiments and analysed using Student's _t_‐test.
Source data are available online for this figure.
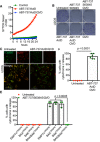
Figure 2. MOMP‐induced mtDNA release initiates a cGAS‐STING‐dependent type I interferon response
- SVEC cells treated with 10 μM ABT‐737 and 10 μM S63845 ± 20 μM qVD‐OPh. Cell viability was analysed using an IncuCyte live‐cell imager and SYTOX Green exclusion. Data are expressed as mean ± SEM, representative of two independent experiments.
- Airyscan images of SVEC cells treated with 10 μM ABT‐737, 10 μM S63845 and 20 μM qVD‐OPh for 3 h, immunostained with anti‐TOM20 and anti‐DNA antibodies. Scale bar = 10 μm. Representative images from three independent experiments.
- Ifnb1, Irf7 and Oasl1 mRNA expression in SVEC cells treated with 10 μM ABT‐737, 10 μM S63845 and 20 μM qVD‐OPh for 3 h. Data are representative of three independent experiments.
- Ifnb1 mRNA expression in SVEC cells treated with 10 μM ABT‐737, 10 μM S63845 ± 20 μM qVD‐OPh for 2 h. Data are representative of two independent experiments.
- STING expression in CRISPR‐Cas9‐mediated STING‐deleted SVEC cells using three independent sgRNA sequences.
- Ifnb1 mRNA expression in STING CRISPR‐Cas9‐deleted SVEC cells treated with 10 μM ABT‐737, 10 μM S63845 and 20 μM qVD‐OPh for 3 h. Data are representative of two independent experiments.
- BAX and BAK expression in SVEC cells harbouring CRISPR‐Cas9‐mediated deletion of BAX, BAK or BAX/BAK.
- Ifnb1 mRNA expression in BAX, BAK or BAX/BAK CRISPR‐Cas9‐deleted SVEC cells treated with 10 μM ABT‐737, 10 μM S63845 and 20 μM qVD‐OPh for 3 h. Data are representative of two independent experiments.
Source data are available online for this figure.
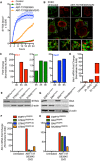
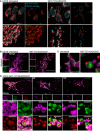
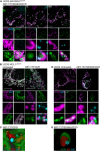
Figure 3. mtDNA extrusion occurs through expanding outer membrane pores
- Airyscan images of U2OS cells stably expressing JF646‐MOM (magenta) treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh for 3 h, immunostained with anti‐DNA and anti‐TFAM antibodies. Scale bar = 10 μm. Representative images from two independent experiments.
- Live‐cell Airyscan imaging of U2OS cells stably expressing JF646‐MOM (magenta) and Omi‐mCherry (red) and transiently expressing TFAM‐mClover (green). Cells were treated with 10 μM ABT‐737, 10 μM S63845 and 20 μM qVD‐OPh at t = 0. Arrows denote mitochondria which show TFAM release. Scale bar = 10 μm. See Video EV4. Numbers indicate time in minutes.
- Zoom of a single mitochondrion from Fig 3B and Video EV4. Scale bar = 1 μm. Numbers indicate time in minutes.
- Loss of Omi intensity as assessed by standard deviation of the Omi signal across the cell and pore size plotted against time. Data are from three mitochondria (pore size) or three cells (Omi release) from two independent experiments and are expressed as mean ± SEM.
Source data are available online for this figure.
Figure EV2. Kinetic analysis of mitochondrial TFAM release
- Airyscan image of U2OS cells transfected with TFAM‐mScarlet and immunostained with anti‐TOM20 and anti‐DNA antibodies. Scale bar = 10 μm. Representative images from two independent experiments.
- Live‐cell Airyscan imaging of U2OS cells stably expressing JF646‐MOM and transiently expressing TFAM‐mClover. Cells were treated with 10 μM ABT‐737 and 20 μM qVD‐OPh at t = 0. Scale bar = 10 μm. See [Link], [Link]. Numbers indicate time in minutes. Arrowheads indicate instances of released TFAM.
- Quantification of mitochondria which do or do not release TFAM as assessed by eye from live‐cell Airyscan imaging, including Fig 3B and Video EV4. Data are from four cells across two independent experiments.
- Time of Omi release and TFAM released were assessed by eye from live‐cell Airyscan imaging, including Fig 3B and Video EV4. Data are from four cells across two independent experiments.
Source data are available online for this figure.
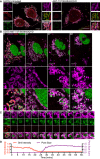
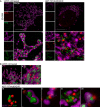
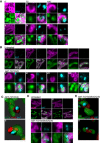
Figure 4. Mitochondrial inner membrane permeabilisation allows mtDNA release into the cytosol
- Maximum intensity projection of Airyscan _z_‐stack data of U2OS cells treated with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh for 3 h. Cells were immunostained with anti‐AIF (IMM) and anti‐DNA antibodies. _Z_‐stack data were 3D‐rendered in Imaris and mtDNA nucleoid inside and outside the AIF signal were visualised. Scale bar = 5 μm. See [Link], [Link]. Representative images from three independent experiments.
- Airyscan images of U2OS cells stably expressing JF646‐MOM (magenta) treated with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh for 3 h and immunostained with anti‐DNA (red) and anti‐AIF (IMM, green) antibodies. Scale bar = 10 μm. Representative images from three independent experiments.
- Imaris 3D‐renderings of MOM (magenta), AIF (green) and DNA (red) from U2OS cells treated as in (B).
- U2OS cells stably expressing JF646‐MOM (magenta) and transiently expressing TFAM‐mScarlet (red) and AIF (1‐90)‐mClover (green) live‐cell imaged by Airyscan. Cells were treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh at t = 0. Scale bar = 10 and 2 μm for zooms. See Video EV7. Representative images from two independent experiments. Numbers indicate time in minutes. Arrowheads indicate instances of released TFAM.
Figure EV3. 3D‐SIM analysis of mtDNA release
- 3D‐SIM images of U2OS cells treated with 10 μM ABT‐737, 1 μM ActD, and 20 μM qVD‐OPh for 3 h. Prior to treatment cells were labelled with JF646 (to label SNAP‐MOM) and post‐fixation were immunostained for IMM (AIF) and DNA. Scale bar = 10 μm. Representative images from three independent experiments.
- Further examples of untreated (i–ii) and treated (iii–iv) U2OS cells 3D‐rendered for MOM (magenta), AIF (IMM, green), and DNA (red). (v–vii) show only the MOM 3D‐rendered with AIF and DNA in captured form.
Source data are available online for this figure.
Figure EV4. mtDNA release is independent of mitochondrial fission and the mitochondrial permeability transition pore
- Expression of Drp1 protein in Wt and _Drp1_‐deleted MEFs.
- IncuCyte live‐imaging of SYTOX Green exclusion in Wt and _Drp1_‐deleted cells treated with 10 μM ABT‐737 and 1 μM ActD to assess cell viability. Data are expressed as mean ± SEM, representative of two independent experiments and have been normalised to starting confluency.
- Expression of CypD protein in Wt and _CypD_‐deleted MEFs.
- U2OS cells loaded with calcein‐AM and CoCl2 and imaged every 30 s for 20 min to show absence of photobleaching. Scale bar = 10 μm. Numbers indicate time in minutes.
- Images from time‐lapse live‐cell imaging of U2OS cells loaded with calcein‐AM and CoCl2 and treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh in the presence of 25 μM CsA at t = 0. Scale bar = 10 μm. See Video EV9. Representative images from two independent experiments. Numbers indicate time in minutes.
Figure 5. mtDNA release is independent of fission and the mitochondrial permeability transition pore
- Maximum intensity projection of z_‐stack Airyscan images of Drp1_fl/fl MEFs with induced Drp1 deletion by AdCre lentiviral particles. Cells were treated with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh for 3 h and immunostained with anti‐TOM20 (MOM) and anti‐DNA antibodies. Zooms show Imaris 3D reconstructions of surface (MOM, TOM20) and spots (DNA) to show the extent of mtDNA release. Scale bar = 10 μm. Representative images from two independent experiments.
- Quantification of mtDNA release per cell in Wt or _Drp1_‐deleted cells. Data are from two independent experiments and are expressed as mean ± SD. Data were analysed by Student's _t_‐test.
- U2OS MCL1CRISPR cells were pre‐treated with 10 μM CCCP and 20 μM qVD‐OPh for 30 min to induce mitochondrial fragmentation after which treatment was changed to 10 μM ABT‐737 and 20 μM qVD‐OPh. After 3 h, cells were fixed and immunostained with anti‐TOM20 and anti‐DNA antibodies. Zooms show highlighted areas. Scale bar = 10 μm. Representative images from three independent experiments.
- Images from time‐lapse live‐cell imaging of U2OS cells loaded with calcein‐AM and CoCl2 and treated with 10 μM ABT‐737/S63845/qVD‐OPh at t = 0. Scale bar = 10 μm. See Video EV8. Representative images from three independent experiments. Numbers indicate time in minutes.
- Quantification of calcein release from mitochondria relative to Omi release. Data are from three independent experiments.
- Maximum intensity projections of _z_‐stack Airyscan images of Wt and CypD −/− MEFs. Cells were treated with 10 μM ABT‐737, 1 μM ActD and 20 μM qVD‐OPh for 3 h and immunostained for TOM20 (MOM) and DNA. Scale bar = 10 μm. Representative images from two independent experiments.
- Quantification of mtDNA release per cell in Wt or _CypD_‐deleted cells. Data are from two independent experiments and are expressed as mean ± SD. Data were analysed by Student's _t_‐test.
Source data are available online for this figure.
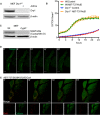
Figure 6. Mitochondrial inner membrane and mtDNA are extruded through BAX pores on the mitochondrial outer membrane
- Live‐cell Airyscan images of U2OS BAX/BAK CRISPR‐Cas9‐deleted cells stably expressing JF646‐MOM (magenta) and mCherry‐BAX (cyan) and transiently expressing TFAM‐mClover (green). Cells were treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh at t = 0. Scale bar = 10 μm. See Video EV10. Numbers indicate time in minutes.
- Airyscan images of U2OS MCL1CRISPR cells stably expressing JF646‐MOM (magenta) and transiently expressing TFAM‐mClover (green), treated with 10 μM ABT‐737 and 20 μM qVD‐OPh for 3 h, immunostained with anti‐active BAX (6A7, cyan). Images are maximum intensity projections of _z_‐stack data. Scale bar = 10 μm. Representative images from three independent experiments.
- Imaris 3D‐rendering of U2OS cells as in (B) showing MOM (green), TFAM (red) and active BAX (cyan).
- U2OS cells stably expressing JF646‐MOM (magenta) treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh for 3 h. Cells were immunostained with antibodies for active BAX (6A7, cyan) and AIF (IMM, green). Images are maximum intensity projections of _z_‐stack data. Scale bar = 10 μm. Representative images from three independent experiments.
- Imaris 3D‐rendering of U2OS cells as in (D) showing MOM (green), AIF (red) and active BAX (cyan).
Figure EV5. Mitochondrial inner membrane and mtDNA are extruded through BAX pores on the mitochondrial outer membrane
- Further examples of stills from live‐cell imaging of U2OS BAX/BAK CRISPR cells expressing JF646‐MOM (magenta) with BAX foci (cyan) and TFAM extrusion (green) from Fig 6A and Video EV10. Cells were treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh at t = 0. Numbers indicate time in minutes.
- Further examples of TFAM extrusion through BAX pores from Fig 6B of U2OS cells stably expressing JF646‐MOM (magenta), transiently expressing TFAM‐mClover (green) and immunostained for active BAX (6A7, cyan). Cells were treated with 10 μM ABT‐737 and 20 μM qVD‐OPh for 3 h.
- Further examples of Imaris 3D‐renderings from Fig 6B of U2OS cells stably expressing JF646‐MOM (green), transiently expressing TFAM‐mClover (red) and immunostained for active BAX (6A7, cyan). Cells were treated with 10 μM ABT‐737 and 20 μM qVD‐OPh for 3 h.
- Further examples of IMM extrusion through BAX pores from Fig 6D of U2OS cells stably expressing JF646‐MOM (magenta) and immunostained for AIF (IMM, green) and active BAX (6A7, cyan). Cells were treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh for 3 h.
- Further examples of Imaris 3D‐renderings from Fig 6D of U2OS cells stably expressing JF646‐MOM (green) and immunostained for AIF (IMM, red) and active BAX (6A7, cyan). Cells were treated with 10 μM ABT‐737, 2 μM S63845 and 20 μM qVD‐OPh for 3 h.
Comment in
- MIM through MOM: the awakening of Bax and Bak pores.
Cosentino K, García-Sáez AJ. Cosentino K, et al. EMBO J. 2018 Sep 3;37(17):e100340. doi: 10.15252/embj.2018100340. Epub 2018 Aug 22. EMBO J. 2018. PMID: 30135068 Free PMC article.
Similar articles
- Role of released mitochondrial DNA in acute lung injury.
Long G, Gong R, Wang Q, Zhang D, Huang C. Long G, et al. Front Immunol. 2022 Aug 18;13:973089. doi: 10.3389/fimmu.2022.973089. eCollection 2022. Front Immunol. 2022. PMID: 36059472 Free PMC article. Review. - BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis.
McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V, Geoghegan ND, Chappaz S, Davidson S, San Chin H, Lane RM, Dramicanin M, Saunders TL, Sugiana C, Lessene R, Osellame LD, Chew TL, Dewson G, Lazarou M, Ramm G, Lessene G, Ryan MT, Rogers KL, van Delft MF, Kile BT. McArthur K, et al. Science. 2018 Feb 23;359(6378):eaao6047. doi: 10.1126/science.aao6047. Science. 2018. PMID: 29472455 - Mitochondrial DNA leakage induces odontoblast inflammation via the cGAS-STING pathway.
Zhou L, Zhang YF, Yang FH, Mao HQ, Chen Z, Zhang L. Zhou L, et al. Cell Commun Signal. 2021 May 20;19(1):58. doi: 10.1186/s12964-021-00738-7. Cell Commun Signal. 2021. PMID: 34016129 Free PMC article. - Mitochondria and Inflammation: Cell Death Heats Up.
Vringer E, Tait SWG. Vringer E, et al. Front Cell Dev Biol. 2019 Jun 27;7:100. doi: 10.3389/fcell.2019.00100. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31316979 Free PMC article. Review.
Cited by
- Too much death can kill you: inhibiting intrinsic apoptosis to treat disease.
Li K, van Delft MF, Dewson G. Li K, et al. EMBO J. 2021 Jul 15;40(14):e107341. doi: 10.15252/embj.2020107341. Epub 2021 May 26. EMBO J. 2021. PMID: 34037273 Free PMC article. Review. - Role of released mitochondrial DNA in acute lung injury.
Long G, Gong R, Wang Q, Zhang D, Huang C. Long G, et al. Front Immunol. 2022 Aug 18;13:973089. doi: 10.3389/fimmu.2022.973089. eCollection 2022. Front Immunol. 2022. PMID: 36059472 Free PMC article. Review. - Mitochondrion-based organellar therapies for central nervous system diseases.
Zhao M, Wang J, Zhu S, Wang M, Chen C, Wang L, Liu J. Zhao M, et al. Cell Commun Signal. 2024 Oct 10;22(1):487. doi: 10.1186/s12964-024-01843-z. Cell Commun Signal. 2024. PMID: 39390521 Free PMC article. Review. - Role of Mitochondrial Nucleic Acid Sensing Pathways in Health and Patho-Physiology.
Chowdhury A, Witte S, Aich A. Chowdhury A, et al. Front Cell Dev Biol. 2022 Feb 11;10:796066. doi: 10.3389/fcell.2022.796066. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35223833 Free PMC article. Review. - Cigarette smoke induces mitochondrial DNA damage and activates cGAS-STING pathway: application to a biomarker for atherosclerosis.
Ueda K, Sakai C, Ishida T, Morita K, Kobayashi Y, Horikoshi Y, Baba A, Okazaki Y, Yoshizumi M, Tashiro S, Ishida M. Ueda K, et al. Clin Sci (Lond). 2023 Jan 31;137(2):163-180. doi: 10.1042/CS20220525. Clin Sci (Lond). 2023. PMID: 36598778 Free PMC article.
References
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD (2005) Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434: 658–662 - PubMed
- Bonora M, Morganti C, Morciano G, Giorgi C, Wieckowski MR, Pinton P (2016) Comprehensive analysis of mitochondrial permeability transition pore activity in living cells using fluorescence‐imaging‐based techniques. Nat Protoc 11: 1067–1080 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/K008374/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 20145/CRUK_/Cancer Research UK/United Kingdom
- 23983/CRUK_/Cancer Research UK/United Kingdom
- MR/L016354/1/MRC_/Medical Research Council/United Kingdom
- C40872/A20145/CRUK_/Cancer Research UK/United Kingdom
- R01 GM123266/GM/NIGMS NIH HHS/United States
- 22903/CRUK_/Cancer Research UK/United Kingdom
- BB/K017314/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 15816/CRUK_/Cancer Research UK/United Kingdom
- MR/K01563X/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials