Mettl14 Is Essential for Epitranscriptomic Regulation of Striatal Function and Learning - PubMed (original) (raw)
. 2018 Jul 25;99(2):283-292.e5.
doi: 10.1016/j.neuron.2018.06.007. Epub 2018 Jun 28.
Lou Dore 2, Hailing Shi 2, Meera J Patel 3, Lee O Vaasjo 1, Meghana N Rao 1, Kai Chen 2, Zhike Lu 2, Yangtian Yi 1, Wanhao Chi 4, Chuan He 5, Xiaoxi Zhuang 6
Affiliations
- PMID: 30056831
- PMCID: PMC6082022
- DOI: 10.1016/j.neuron.2018.06.007
Mettl14 Is Essential for Epitranscriptomic Regulation of Striatal Function and Learning
Jessica L Koranda et al. Neuron. 2018.
Abstract
N6-methyladenosine (m6A) regulates mRNA metabolism and translation, serving as an important source of post-transcriptional regulation. To date, the functional consequences of m6A deficiency within the adult brain have not been determined. To achieve m6A deficiency, we deleted Mettl14, an essential component of the m6A methyltransferase complex, in two related yet discrete mouse neuronal populations: striatonigral and striatopallidal. Mettl14 deletion reduced striatal m6A levels without altering cell numbers or morphology. Transcriptome-wide profiling of m6A-modified mRNAs in Mettl14-deleted striatum revealed downregulation of similar striatal mRNAs encoding neuron- and synapse-specific proteins in both neuronal types, but striatonigral and striatopallidal identity genes were uniquely downregulated in each respective manipulation. Upregulated mRNA species encoded non-neuron-specific proteins. These changes increased neuronal excitability, reduced spike frequency adaptation, and profoundly impaired striatal-mediated behaviors. Using viral-mediated, neuron-specific striatal Mettl14 deletion in adult mice, we further confirmed the significance of m6A in maintaining normal striatal function in the adult mouse.
Keywords: Mettl14; N(6)-methyladenosine; epitranscriptome; m(6)A; mRNA methylation; striatal learning; striatonigral neurons; striatopallidal neurons; translational regulation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures
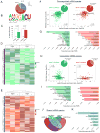
Figure 1. Mettl14 deletion in striatonigral or striatopallidal neurons alters striatal epitranscriptome
A) Transcriptome-wide distribution of m6A peaks of control samples. Number and percent of m6A peaks are shown. B) Consensus motif of m6A sites identified in m6A peaks of control samples. C) LC-MS/MS showing knockdown of striatal m6A levels. Data normalized to control percent methylated adenosine. D & E) Clustering and heat maps showing mRNA levels in three experimental and control mice following striatonigral (D) or striatopallidal (E) Mettl14 deletion. Relative mRNA expression level is represented in color. Only genes with log2 of enrichment fold of m6A peaks m6A-seq > 1 in controls and significant changes (p<0.05) in mRNA expression levels are shown. **F)** Correlation between (1) the fold change (FC) of RNA abundance in experimental relative to control mice and (2) the log2 ratio of m6A enrichment (experimental/control) in significantly downregulated mRNAs. _P_ values were calculated from a Pearson’s product-moment correlation. _left:_ D1R-fM14exp, _right:_ D2R-fM14exp. _Insets:_ Pie chart showing significantly altered m6A-containing and non-m6A-containing mRNA species. **G)** GO analysis of mRNAs with RPKM ratio< 1 and _p_ <0.05. _left:_ D1R-fM14exp, _right:_ D2R-fM14exp. Number of genes in each category shown in parentheses. **H)** Correlation analysis as described for (F) using significantly upregulated mRNAs. _left:_ D1R-fM14exp, _right:_ D2R-fM14exp. _Insets:_ Pie chart showing significantly altered m6A-containing and non-m6A-containing mRNA species. **I)** GO analysis with FC > 1 and p < 0.05. left: D1R-fM14exp, right: D2R-fM14exp. Number of genes in each category shown in parentheses. J) Venn diagram showing overlap of genes with significant changes in mRNA expression in both D1R-fM14exp and D2R-fM14exp mice. K) GO analysis of genes with significantly downregulated mRNA species in both D1R-fM14exp and D2R-fM14exp mice. Number of genes in each category shown in parentheses. See also Fig. S1 and Table S1.
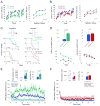
Figure 2. Lack of Mettl14 in striatal subpopulations alters striatal-mediated behavior
A and B) Latency to fall from the accelerating rotarod in 12 week old mice. Left: Mice were given 5 trials per day. Right: Each data point represents the average of 5 trials per training session. (A) D1R-fM14ctrl, n = 7, D1R-fM14exp, n =8; (B) D2R-fM14ctrl, n = 17, D2R-fM14exp, n = 14. C) Response learning acquisition (left) and reversal learning (right) in mice with Mettl14 deleted in striatonigral (top) and striatopallidal (bottom) neurons using water-cross maze. Survival plots show percent mice remaining below learning criterion. D1R-fM14exp: n = 6; D1R-fM14ctrl: n =5. D2R-fM14exp: n=6, D2R-fM14ctrl: n=7. D) Latency to reach a visible platform in the water maze for entire session (top) and across 5 trials (bottom). Left: D1R-fM14exp, n = 6; D1R-fM14ctrl, n =5. Right: D2R-fM14exp: n=6, D2R-fM14ctrl: n=7. E) Locomotor response of D1R-fM14exp and control mice to a single injection of the D1R agonist SKF82197 (8.0 mg/kg) across 1 min bins (bottom). Total ambulatory distance across 60 minutes (top, left) and % change in locomotion following SKF81297 injection relative to saline (top, right). D1R-fM14exp: n = 8; D1R-fM14ctrl: n =7. F) Locomotor response of D2R-fM14exp and control mice to a single injection of the D2R antagonist eticlopride (0.16 mg/kg) across 1 min bins (bottom). Total ambulatory distance across 60 minutes (top, left) and % change in locomotion following eticlopride injection relative to saline (top, right). D2R-fM14exp: n = 14; D2R-fM14ctrl: n =17. All data expressed as mean ± S.E.M.
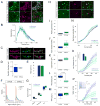
Figure 3. Morphological and physiological analysis of striatonigral neurons lacking Mettl14 A)
Representative immunohistochemistry showing Mettl14 deletion in mCherry labelled D1R-expressing striatal neurons. B) Sholl analysis assessing dendritic branching as a function of distance from soma. C) Representative dendrites from showing sections of an mCherry-filled dendrite (left), reconstruction of dendrites (middle), and overlay of the reconstruction on the actual dendrite (right). D) Quantification of dendritic diameter. E) Total spine number. F) Frequency plots of spine volume. G) Average number of spines according to classification. D1R-fM14ctrl: n = 35 dendrites, 5 mice. D1R-fM14exp: n = 21 dendrites, 3 mice. H) Representative cells showing presence of METTL14 in a recorded control cell and the absence of METTL14 in a recorded experimental cell filled with biocytin. Intrinsic membrane properties of D1R-fM14exp mice, including (I) resting membrane potential, (J) input resistance, (K) capacitance, (L) rheobase and (M) paired pulse ratio (PPR). D1R-fM14exp: n = 15 cells, 4 mice. D1R-fM14ctrl: n = 23 cells, 6 mice. N) Membrane responses to current injection. D1R-fM14exp: n = 11 cells, 4 mice. D1R-fM14ctrl: n = 18 cells, 6 mice. O) Instantaneous firing rate responses to current injection. Inset: Average current to elicit train of action potentials. Cells that did not generate a train of spikes were not considered. D1R-fM14exp: n = 11 cells, 4 mice. D1R-fM14ctrl: n = 15 cells, 6 mice. P) Average spike number in response to current injection. Inset: Example of a trace elicited from cells with an ISI of ~14 Hz following 325 pA current injection. D1R-fM14exp: n = 11 cells, 4 mice. D1R-fM14ctrl: n = 15 cells, 6 mice. Box plot whiskers show min to max values. Black points indicate outliers. Data shows average across each mouse ± S.E.M. See also Table S2.
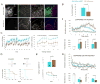
Figure 4. Viral-mediated deletion of Mettl14 in the dorsal striatum of adult mice alters striatal learning and response to dopaminergic drugs
A) Representative pictures showing deletion of METTL14 in adult dorsal striatal neurons following injection of an AAV expressing either eGFP (fM14;Syn-eGFP) or Cre recombinase (fM14;Syn-Cre) under control of the neuronal promotor synapsin (Syn). Yellow dotted line denotes approximate area of viral expression. B) LC-MS/MS showing reduced levels of m6A mRNA species. Data is normalized to control percent methylated adenosine relative to total adenosine. C) Latency to fall from the accelerating rotarod at 4 and 8 weeks post-injection in individual trials (left) or across whole session (right). N=9 for each group. D) Response learning acquisition (left) and reversal learning (right) in the water-cross maze. Survival graphs show mice reaching criteria for each phase. N=9 for each group. E) Latency to reach a visible platform in the water maze across all 5 trials (top) and for each individual trial (bottom). N=9 for each group. F) Locomotor response to a single injection of D1R agonist SKF82197 (8.0 mg/kg, SKF) across 2 min bins (top). Total ambulatory distance across 60 minutes (bottom, left) and % change in locomotion following SKF81297 injection relative to saline (bottom, right). Syn-eGFP: n=10, Syn-Cre: n=8. G) Locomotor response to a single injection of D2R antagonist eticlopride (0.16mg/kg, etic) across 2 min bins (top). Total ambulatory distance across 60 minutes (bottom, left) and % change in locomotion following eticlopride injection relative to saline (bottom, right). Syn-eGFP: n=9; Syn-Cre n=9. Data shows average across each mouse ± S.E.M. See also Fig. S2.
Comment in
- Epitranscriptomes in the Adult Mammalian Brain: Dynamic Changes Regulate Behavior.
Yoon KJ, Ming GL, Song H. Yoon KJ, et al. Neuron. 2018 Jul 25;99(2):243-245. doi: 10.1016/j.neuron.2018.07.019. Neuron. 2018. PMID: 30048610 Free PMC article.
Similar articles
- FACS array profiling identifies Ecto-5' nucleotidase as a striatopallidal neuron-specific gene involved in striatal-dependent learning.
Ena SL, De Backer JF, Schiffmann SN, de Kerchove d'Exaerde A. Ena SL, et al. J Neurosci. 2013 May 15;33(20):8794-809. doi: 10.1523/JNEUROSCI.2989-12.2013. J Neurosci. 2013. PMID: 23678122 Free PMC article. - Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function.
Teng Y, Liu Z, Chen X, Liu Y, Geng F, Le W, Jiang H, Yang L. Teng Y, et al. J Cell Mol Med. 2021 Sep;25(17):8567-8572. doi: 10.1111/jcmm.16740. Epub 2021 Jul 21. J Cell Mol Med. 2021. PMID: 34288397 Free PMC article. - YTHDF1 mediates translational control by m6A mRNA methylation in adaptation to environmental challenges.
Shi Z, Wen K, Zou Z, Fu W, Guo K, Sammudin NH, Ruan X, Sullere S, Wang S, Zhang X, Thinakaran G, He C, Zhuang X. Shi Z, et al. bioRxiv [Preprint]. 2024 Aug 9:2024.08.07.607063. doi: 10.1101/2024.08.07.607063. bioRxiv. 2024. PMID: 39149343 Free PMC article. Preprint. - N (6)-Methyladenosine (m(6)A) Methylation in mRNA with A Dynamic and Reversible Epigenetic Modification.
Wu R, Jiang D, Wang Y, Wang X. Wu R, et al. Mol Biotechnol. 2016 Jul;58(7):450-9. doi: 10.1007/s12033-016-9947-9. Mol Biotechnol. 2016. PMID: 27179969 Review. - Integrating neurotransmission in striatal medium spiny neurons.
Girault JA. Girault JA. Adv Exp Med Biol. 2012;970:407-29. doi: 10.1007/978-3-7091-0932-8_18. Adv Exp Med Biol. 2012. PMID: 22351066 Review.
Cited by
- METTL14-mediated upregulation of lncRNA HOTAIR represses PP1α expression by promoting H3K4me1 demethylation in oxycodone-treated mice.
Liu TC, Li HX, Wan YX, Shi G, Zhao YP, Liu YF, Fan XY. Liu TC, et al. CNS Neurosci Ther. 2024 Jul;30(7):e14830. doi: 10.1111/cns.14830. CNS Neurosci Ther. 2024. PMID: 39046182 Free PMC article. - Potential role of N6-methyladenosine modification in the development of Parkinson's disease.
Zhou J, Han Y, Hou R. Zhou J, et al. Front Cell Dev Biol. 2023 Dec 13;11:1321995. doi: 10.3389/fcell.2023.1321995. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38155838 Free PMC article. Review. - RNA N6-Methyladenosine and the Regulation of RNA Localization and Function in the Brain.
Madugalle SU, Meyer K, Wang DO, Bredy TW. Madugalle SU, et al. Trends Neurosci. 2020 Dec;43(12):1011-1023. doi: 10.1016/j.tins.2020.09.005. Epub 2020 Oct 8. Trends Neurosci. 2020. PMID: 33041062 Free PMC article. Review. - The m6A Dynamics of Profilin in Neurogenesis.
Rockwell AL, Hongay CF. Rockwell AL, et al. Front Genet. 2019 Nov 12;10:987. doi: 10.3389/fgene.2019.00987. eCollection 2019. Front Genet. 2019. PMID: 31798620 Free PMC article. Review. - High glucose induces tau hyperphosphorylation in hippocampal neurons via inhibition of ALKBH5-mediated Dgkh m6A demethylation: a potential mechanism for diabetic cognitive dysfunction.
Qu M, Zuo L, Zhang M, Cheng P, Guo Z, Yang J, Li C, Wu J. Qu M, et al. Cell Death Dis. 2023 Jun 29;14(6):385. doi: 10.1038/s41419-023-05909-7. Cell Death Dis. 2023. PMID: 37385994 Free PMC article.
References
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–89. - PubMed
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RM1 HG008935/HG/NHGRI NIH HHS/United States
- R01 HG008688/HG/NHGRI NIH HHS/United States
- T32 MH020065/MH/NIMH NIH HHS/United States
- T32 DA043469/DA/NIDA NIH HHS/United States
- R01 DA043361/DA/NIDA NIH HHS/United States
- R01 GM113194/GM/NIGMS NIH HHS/United States
- P01 NS097206/NS/NINDS NIH HHS/United States
- R21 NS103159/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous