TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells - PubMed (original) (raw)
TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells
Dana Elster et al. Nat Commun. 2018.
Erratum in
- Author Correction: TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells.
Elster D, Tollot M, Schlegelmilch K, Ori A, Rosenwald A, Sahai E, von Eyss B. Elster D, et al. Nat Commun. 2018 Sep 12;9(1):3781. doi: 10.1038/s41467-018-06266-2. Nat Commun. 2018. PMID: 30209298 Free PMC article.
Abstract
Yes-associated protein (YAP), the downstream transducer of the Hippo pathway, is a key regulator of organ size, differentiation and tumorigenesis. To uncover Hippo-independent YAP regulators, we performed a genome-wide CRISPR screen that identifies the transcriptional repressor protein Trichorhinophalangeal Syndrome 1 (TRPS1) as a potent repressor of YAP-dependent transactivation. We show that TRPS1 globally regulates YAP-dependent transcription by binding to a large set of joint genomic sites, mainly enhancers. TRPS1 represses YAP-dependent function by recruiting a spectrum of corepressor complexes to joint sites. Loss of TRPS1 leads to activation of enhancers due to increased H3K27 acetylation and an altered promoter-enhancer interaction landscape. TRPS1 is commonly amplified in breast cancer, which suggests that restrained YAP activity favours tumour growth. High TRPS1 activity is associated with decreased YAP activity and leads to decreased frequency of tumour-infiltrating immune cells. Our study uncovers TRPS1 as an epigenetic regulator of YAP activity in breast cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1
Identification of the YAP modulator TRPS1 using a genome-wide CRISPR screen. a Schematic of the YAP activity sensor system. The sensor cell line harbours a doxycycline inducible Strep-YAP5SA allele and a turboRFP (red fluorescent protein) reporter under the control of a CTGF promoter fragment containing TEAD-binding sites. b Western blot for YAP and CTGF in sensor cells treated with doxycycline (DOX) or ethanol (EtOH). Vinculin serves as loading control. c qRT-PCR analysis of the sensor cell line for the YAP target genes CTGF and ANKRD1. The cells were treated with doxycycline (DOX) or ethanol (EtOH). The chart summarizes three biological replicates. Error bars represent s.e.m. d Flow cytometry for RFP in the sensor cell line after treatment with doxycycline (DOX) or ethanol (EtOH), respectively. e Schematic of the CRISPR screening strategy. MCF10A sensor cells were infected with the genome-wide lentiviral GeCKO v2 CRISPR library. After doxycycline (DOX) treatment, cells were sorted into two subpopulations, “high” or “low”, representing 1% of the cells with highest or lowest RFP signal, respectively. Both populations were then analyzed by deep sequencing to determine the frequency of each sgRNA. f Plots for the distribution of _P_-values in the low vs. unsorted (left) and high vs. unsorted (right) cells, respectively, after analysis by the redundant siRNA activity (RSA) algorithm. g Workflow for the validation of candidates from the CRISPR screen. h qRT-PCR analysis of ANKRD1 expression in the doxycycline-treated sensor cell line transfected with siCtrl or siRNA targeting candidate YAP modulators. The cells were treated with doxycycline (+DOX) to induce YAP 5SA expression or ethanol (EtOH) as a control. Data presented are means from technical triplicates and error bars represent s.d. i Schematic of the TRPS1 protein
Fig. 2
TRPS1 represses YAP target genes. a Western blot for YAP from MCF7 i5SA (pInducer-Strep YAP 5SA MCF7) cells after 14-h induction by doxycycline (DOX) or ethanol (EtOH), respectively. Vinculin serves as loading control. b MA plot for RNA-Sequencing from MCF7 i5SA cells after doxycycline (DOX) or ethanol (EtOH) treatment for 14 h. The Log2 fold change (Log2FC) is plotted against the abundance of transcripts given as number of reads per kilobase per million mapped reads (RPKM). YAP-induced genes are drawn as blue dots. c Western blot for TRPS1 and YAP after infection of MCF7 cells with three different TRPS1 shRNAs (shTRPS1 #1–3) or a non-targeting control (shNTC). Vinculin serves as loading control. d Gene set enrichment analysis (GSEA) for a set of 497 YAP-induced genes in RNA-Sequencing from TRPS1-depleted MCF7 cells. NES normalized enrichment score. e MA plot for RNA-Sequencing from MCF7 cells infected with three different TRPS1 shRNAs or a non-targeting control (shNTC). The Log2 fold change (Log2FC) is plotted against the abundance of transcripts given as number of reads per kilobase per million mapped reads (RPKM). YAP-induced genes are drawn as blue dots. Red dots label established YAP targets. f Violin Plot showing the differential regulation of YAP-induced genes after TRPS1 depletion by shTRPS1.The white point indicates the median. Two-sided Wilcox test. g qRT-PCR analysis of several YAP target genes in MCF7 i5SA cells after induction of YAP 5SA expression by doxycycline (DOX) or ethanol control (EtOH). Data presented are means from three biological replicates (n = 3) and error bars represent s.e.m; *P < 0.05; **P < 0.01; ***P < 0.001; Student’s _t_-test. h qRT-PCR analysis of the YAP target genes from g in MCF7 cells infected with three different TRPS1 shRNAs or a non-targeting control (shNTC). Data presented are means from technical triplicates per shRNA and error bars represent s.d.; *P < 0.05; **P < 0.01; ***P < 0.001; Student’s _t_-test
Fig. 3
TRPS1 and YAP/TEAD bind to an overlapping set of genomic sites. a Heatmaps of ChIP-Seq data from MCF7 cells showing the occupancy of TRPS1, TEAD1 and YAP at all RefSeq transcriptional start sites (TSSs). The heatmap was sorted according to TRPS1 binding. b Heatmaps of ChIP-Seq data from MCF7 cells showing the occupancy of TRPS1, TEAD1 and YAP at all enhancer regions. The heatmap was sorted according to TRPS1 binding. The enhancer-specific chromatin marks were taken from a previously published data set. The same contrasts for promoters (a) and enhancers (b) were used to demonstrate the differences in binding strength between the two. c Venn diagram showing the numbers of promoters and enhancers, respectively, bound by TRPS1, TEAD1 and YAP in MCF7 cells. d ChIP-Seq density profiles for H3K27ac, YAP and TEAD1 in a ±2.5-kb window surrounding the centre of TRPS1 peaks at enhancer sites. e Venn diagram for the numbers of ATAC-Seq peaks in T47D cells transfected with control siRNAs (siCtrl) or a siRNAs targeting TRPS1, respectively. f Distribution of ATAC-Seq reads with an insert size ≥146 bp (left), or with an insert size <146 bp (right) at enhancers bound by TRPS1 after TRPS1 depletion by siRNAs in T47D cells. _P_-values were calculated using a two-sided Wilcox-test. Rep1 replicate 1, Rep2 replicate 2. Indicated below are _P_-values for ATAC-Seq signals that describe if enhancers bound by TRPS1 are more strongly affected by TRPS1 depletion than enhancers not bound by TRPS1. Two-sided Wilcox-test. g Centrimo analysis for TEAD1 and TEAD3 binding motifs at TRPS1-bound enhancer sites in a 1-kb window. The reads are centred on the respective TRPS1 peak. h Sequencing tracks for ChIP-Seq data and ATAC-Seq data of the TGFB2 locus. The last row shows the Log2 fold change (Log2FC) of TGFB2 expression determined by RNA-Seq in MCF7 and T47D cells after TRPS1 depletion and in MCF7 cells after overexpression of YAP 5SA
Fig. 4
TRPS1 gets recruited to TEAD sites by cooperative binding. a ChIP-Seq tracks for TRPS1, TEAD1, YAP, H3K27ac and H3K4me1 at the VTCN1 Enhancer region. b Motif enrichment analysis (DREME) for GATA4 and GATA1 binding sites in TRPS1 ChIP peaks. c Centrimo analysis for GATA1 and TEAD1 motifs to identify enriched binding sites surrounding the Top 500 TRPS1 ChIP-Seq peaks. d Representative pictures from proximity ligation assay (PLA) experiments in MCF7 cells. The pictures show PLA signals (in green) for YAP and TEAD (positive control, left) or TEAD1 and TRPS1 after treatment with siRNAs targeting TRPS1 or control siRNAs (siCtrl). e Quantification of PLA signals for the TEAD1-TRPS1 interaction in MCF7 cells treated with siTRPS1 or siCtrl, respectively. Values are presented as the number of dots per nucleus for each of the three biological replicates. A horizontal bar indicates the average. For quantification, at least 400 cells per replicate were counted. Student’s _t_-test. f Co-immunoprecipitation experiment from 293T cells co-expressing TRPS1 and the indicated GAL4-tagged TEAD proteins. The lysates were subjected to immunoprecipitations using a TRPS1 antibody or IgG as a control and subsequently analyzed by Western blot. IgG controls and TRPS1 precipitates were analyzed on the same membrane. g Endogenous co-immunoprecipitation experiment from MCF7 WT and KO TRPS1. The lysates were subjected to immunoprecipitations using a TRPS1 antibody and subsequently analyzed by Western blot using a Pan-TEAD antibody; h.c. heavy chain
Fig. 5
TRPS1 recruits corepressor complexes to chromatin. a Scheme depicting the BioID method. Mutant BirA (BirA*) fused to TRPS1 biotinylates all proteins in close proximity to the fusion protein after addition of biotin. b Western blot for BirA*-Flag-TRPS1 and the NLS-BirA*-Flag control from transfected 293T cells. Vinculin serves as loading control. c Heatmap clustering analysis (k-nearest neighbour) for the biological triplicates of the BioID experiment. d Volcano plot for nuclear proteins detected in all three BioID replicates. Log2FC: Log2 fold change; P. adj: adjusted _P_-value. e Western blot for CTBP2, SMRT, HDAC1 in TRPS1 co-immunoprecipitates from 293T cells expressing V5-TRPS1. f qChIP analysis for TRPS1, CTBP2, SMRT and HDAC3 binding at several enhancers and at the BMP7 promoter. The U2 promoter served as a control region. Control IPs were performed with rabbit or mouse IgG, respectively. Error bars represent s.d.
Fig. 6
Loss of TRPS1 alters chromatin structure and long-range interactions. a Pie charts depicting the proportion of enhancer and promoter regions among CTBP2 and HDAC3 peaks. b Venn diagrams showing the overlap of enhancer sites bound by TRPS1 and CTBP2 (left) or TRPS1 and HDAC3 (right). c Western blot for TRPS1 and YAP in MCF7 WT and TRPS1 KO cells. d, e Density profiles for CTBP2 (d) and HDAC3 (e) binding at enhancers bound by TRPS1 in MCF7 and TRPS1 KO cells. f Density profiles for H3K27ac at enhancers bound by TRPS1 in MCF7 WT and TRPS1 KO cells. g _P_-values that describe if enhancers bound by TRPS1 are more strongly affected by TRPS1 deletion than enhancers not bound by TRPS1. Wilcox-test, two-sided. h Venn diagram showing the number of overlapping and unique H3K27ac ChIP peaks in MCF7 WT and TRPS1 KO cells, respectively. i ChIP-Seq tracks for TRPS1, TEAD1, YAP and H3K27ac from MCF7 WT and TRPS1 KO cells at an enhancer at the given genomic location. The annotated enhancer is drawn as a pink rectangle. The 2 kb fragment that was used for the luciferase assay in j is drawn as a black rectangle. j Luciferase activity of the reporter driven by the enhancer from i in MCF7 WT and TRPS1 KO cells co-transfected with the indicated expression vectors. Increasing amounts of a FLAG-YAP 5SA construct and constant amounts of HA-TEAD construct were used. Data presented are derived from three biological replicates and error bars represent s.e.m. Student’s _t_-test. k 4C-Seq interaction pattern of the IGFBP3 TSS in MCF7 (WT) compared to TRPS1 KO (KO) cells. Significance was determined by the w4CSeq analysis package using a one-tailed binomial test and corrected for multiple testing yielding the adjusted _P_-value (P. adj). The HiC interaction map was used from a previously published ENCODE (ENCSR549MGQ) data set. Black and grey boxes mark TADs and a dashed line marks sub-TADs of the left TAD. The locations of TRPS1 ChIP-Seq peaks and annotated enhancers are given in blue and pink, respectively. TAD topologically associated domain
Fig. 7
TRPS1 is overexpressed in breast cancer and is predictive for survival. a Immunohistochemical staining for TRPS1 on tissue sections from human breast cancer patients. Scale bar = 50 μm. b TRPS1 gene amplification level in tumour vs. normal breast tissue from breast cancer patients of the TCGA data set. c Box plot showing TRPS1 mRNA levels in patients stratified according to the level of amplification of the TRPS1 locus. Median: black line; boxes: data points between the first and third quartiles; whiskers: up to 1.5 × interquartile range; points: outliers. Wilcox-test, two-sided. d Kaplan–Meier plot for the survival probability of breast cancer patients that were stratified based on their TRPS1 amplification status. TRPS1 high: patients with TRPS1 amplification status above the third quartile compared to TRPS1 low (remaining patients). Chi-square test. e Analysis of TRPS1 and YAP activity in breast cancer patients (see Methods). The correlation between TRPS1 and YAP activities is given for all patients (all) and for the given breast cancer subtypes. The correlation coefficients (ρ) and the corresponding _P_-values were determined by a Spearman rank correlation test. f Box plots showing the expression of genes repressed by TRPS1 in different breast cancer subtypes. Median: black line; boxes: data points between the first and third quartiles; whiskers: up to 1.5 × interquartile range; points: outliers. Wilcox-test, two-sided
Fig. 8
TRPS1 is needed for efficient tumour growth in vivo. a Western blot for TRPS1 in 4T1 cells infected with two different shRNAs targeting TRPS1 or a non-targeting shRen (Renilla) control, respectively. Serial dilutions (lanes 1–3) of the shRen control are loaded to demonstrate the knockdown efficiency. b Tumour growth of 4T1-derived tumours orthotopically transplanted into the mammary gland of BALB/C mice. Tumour size was monitored after the indicated times of injection. Data are presented as means ± s.e.m. Two-way ANOVA test. c The tumour weight of the 4T1-derived tumours was determined 23 days after transplantation. One-way ANOVA test. d Photos of 4T1-derived tumours 23 days after transplantation. Scale bar = 1 cm. e Representative images of IHC stainings for CD3-positive cells (T cells) in sections of the 4T1-derived tumours. Scale bar = 50 μm. f Quantification of (k). Error bars represent s.e.m. One-way ANOVA test. g Expression of signature gene sets for natural killers (NK), cytotoxic T cells (CD8) and helper T cells (CD4) were sorted based on TRPS1 activity in breast cancer patients. Spearman rank correlation test
Fig. 9
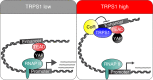
Model of how TRPS1 modulates YAP/TEAD-dependent transcription. See Discussion for details. CoR corepressors
Similar articles
- TRPS1 regulates oestrogen receptor binding and histone acetylation at enhancers.
Serandour AA, Mohammed H, Miremadi A, Mulder KW, Carroll JS. Serandour AA, et al. Oncogene. 2018 Sep;37(39):5281-5291. doi: 10.1038/s41388-018-0312-2. Epub 2018 Jun 12. Oncogene. 2018. PMID: 29895970 Free PMC article. - A Non-canonical Role of YAP/TEAD Is Required for Activation of Estrogen-Regulated Enhancers in Breast Cancer.
Zhu C, Li L, Zhang Z, Bi M, Wang H, Su W, Hernandez K, Liu P, Chen J, Chen M, Huang TH, Chen L, Liu Z. Zhu C, et al. Mol Cell. 2019 Aug 22;75(4):791-806.e8. doi: 10.1016/j.molcel.2019.06.010. Epub 2019 Jul 11. Mol Cell. 2019. PMID: 31303470 Free PMC article. - Nuclear cathepsin D enhances TRPS1 transcriptional repressor function to regulate cell cycle progression and transformation in human breast cancer cells.
Bach AS, Derocq D, Laurent-Matha V, Montcourrier P, Sebti S, Orsetti B, Theillet C, Gongora C, Pattingre S, Ibing E, Roger P, Linares LK, Reinheckel T, Meurice G, Kaiser FJ, Gespach C, Liaudet-Coopman E. Bach AS, et al. Oncotarget. 2015 Sep 29;6(29):28084-103. doi: 10.18632/oncotarget.4394. Oncotarget. 2015. PMID: 26183398 Free PMC article. - Analysis of the function, mechanism and clinical application prospect of TRPS1, a new marker for breast cancer.
He X, Huang H, Liu Y, Li H, Ren H. He X, et al. Gene. 2025 Jan 10;932:148880. doi: 10.1016/j.gene.2024.148880. Epub 2024 Aug 23. Gene. 2025. PMID: 39181273 Review. - The TEAD Family and Its Oncogenic Role in Promoting Tumorigenesis.
Zhou Y, Huang T, Cheng AS, Yu J, Kang W, To KF. Zhou Y, et al. Int J Mol Sci. 2016 Jan 21;17(1):138. doi: 10.3390/ijms17010138. Int J Mol Sci. 2016. PMID: 26805820 Free PMC article. Review.
Cited by
- TRPS1 Is a Lineage-Specific Transcriptional Dependency in Breast Cancer.
Witwicki RM, Ekram MB, Qiu X, Janiszewska M, Shu S, Kwon M, Trinh A, Frias E, Ramadan N, Hoffman G, Yu K, Xie Y, McAllister G, McDonald R, Golji J, Schlabach M, deWeck A, Keen N, Chan HM, Ruddy D, Rejtar T, Sovath S, Silver S, Sellers WR, Jagani Z, Hogarty MD, Roberts C, Brown M, Stegmaier K, Long H, Shivdasani RA, Pellman D, Polyak K. Witwicki RM, et al. Cell Rep. 2018 Oct 30;25(5):1255-1267.e5. doi: 10.1016/j.celrep.2018.10.023. Cell Rep. 2018. PMID: 30380416 Free PMC article. - YAP-independent mechanotransduction drives breast cancer progression.
Lee JY, Chang JK, Dominguez AA, Lee HP, Nam S, Chang J, Varma S, Qi LS, West RB, Chaudhuri O. Lee JY, et al. Nat Commun. 2019 Apr 23;10(1):1848. doi: 10.1038/s41467-019-09755-0. Nat Commun. 2019. PMID: 31015465 Free PMC article. - Emerging Principles in the Transcriptional Control by YAP and TAZ.
Lopez-Hernandez A, Sberna S, Campaner S. Lopez-Hernandez A, et al. Cancers (Basel). 2021 Aug 23;13(16):4242. doi: 10.3390/cancers13164242. Cancers (Basel). 2021. PMID: 34439395 Free PMC article. Review. - Multi-omics profiling of mouse polycystic kidney disease progression at a single cell resolution.
Muto Y, Yoshimura Y, Wu H, Chang-Panesso M, Ledru N, Woodward OM, Outeda P, Cheng T, Mahjoub MR, Watnick TJ, Humphreys BD. Muto Y, et al. bioRxiv [Preprint]. 2024 May 31:2024.05.27.595830. doi: 10.1101/2024.05.27.595830. bioRxiv. 2024. PMID: 38854144 Free PMC article. Updated. Preprint. - Genome-Wide Estrogen Receptor Activity in Breast Cancer.
Farcas AM, Nagarajan S, Cosulich S, Carroll JS. Farcas AM, et al. Endocrinology. 2021 Feb 1;162(2):bqaa224. doi: 10.1210/endocr/bqaa224. Endocrinology. 2021. PMID: 33284960 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases