Transcriptome Analysis of Long Non-coding RNAs and Genes Encoding Paraspeckle Proteins During Human Ovarian Follicle Development - PubMed (original) (raw)
Transcriptome Analysis of Long Non-coding RNAs and Genes Encoding Paraspeckle Proteins During Human Ovarian Follicle Development
Emil H Ernst et al. Front Cell Dev Biol. 2018.
Abstract
Emerging evidence indicated that many long non-coding (lnc)RNAs function in multiple biological processes and dysregulation of their expression can cause diseases. Most regulatory lncRNAs interact with biological macromolecules such as DNA, RNA, and protein. LncRNAs regulate gene expression through epigenetic modification, transcription, and posttranscription, through DNA methylation, histone modification, and chromatin remodeling. Interestingly, differential lncRNA expression profiles in human oocytes and cumulus cells was recently assessed, however, lncRNAs in human follicle development has not previously been described. In this study, transcriptome dynamics in human primordial, primary and small antral follicles were interrogated and revealed information of lncRNA genes. It is known that some lncRNAs form a complex with paraspeckle proteins and therefore, we extended our transcriptional analysis to include genes encoding paraspeckle proteins. Primordial, primary follicles and small antral follicles was isolated using laser capture micro-dissection from ovarian tissue donated by three women having ovarian tissue cryopreserved before chemotherapy. After RN sequencing, a bioinformatic class comparison was performed and primordial, primary and small antral follicles were found to express several lncRNA and genes encoding paraspeckle proteins. Of particular interest, we detected the lncRNAs XIST, NEAT1, NEAT2 (MALAT1), and GAS5. Moreover, we noted a high expression of FUS, TAF15, and EWS components of the paraspeckles, proteins that belong to the FET (previously TET) family of RNA-binding proteins and are implicated in central cellular processes such as regulation of gene expression, maintenance of genomic integrity, and mRNA/microRNA processing. We also interrogated the intra-ovarian localization of the FUS, TAF15, and EWS proteins using immunofluorescence. The presence and the dynamics of genes that encode lncRNA and paraspeckle proteins may suggest that these may mediate functions in the cyclic recruitment and differentiation of human follicles and could participate in biological processes known to be associated with lncRNAs and paraspeckle proteins, such as gene expression control, scaffold formation and epigenetic control through human follicle development. This comprehensive transcriptome analysis of lncRNAs and genes encoding paraspeckle proteins expressed in human follicles could potentially provide biomarkers of oocyte quality for the development of non-invasive tests to identify embryos with high developmental potential.
Keywords: fertility; human follicle; lncRNA; paraspeckle; treatment.
Figures
Figure 1
Schematic illustration of human follicular cells isolated using laser capture microdissection. Please note that the aspect ratio is arbitrary.
Figure 2
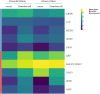
Heatmap representation of selected genes encoding paraspeckle proteins and lncRNA genes. Heatmap of gene expression in oocytes and granulosa cells during the primordial-to-primary follicle transition. The heatmap includes the paraspeckle-encoding genes (blue bar) MEX3C, NONO, FUS_, EWS, TAF15_ transcripts, and the lncRNAs (red bar) NEAT1, MALAT1 (NEAT2), XIST, ZFAS1, GAS5. Color code reflects average FPKM values.
Figure 3
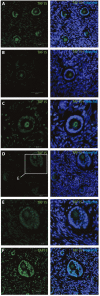
Intra-ovarian distribution of TAF15 in human primordial and primary follicles. This showed detection of the TAF15 protein in both oocyte and granulosa cells of (A) primordial, (B) primary, and (C) secondary, as well as (D–F) small pre-antral/early antral follicles. Hoechst staining identifies the nucleus of cells. Scale bars; 30 μm.
Figure 4
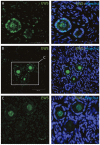
Intra-ovarian distribution of EWS in human primordial and primary follicles (A-C). EWS is present in both oocytes and the surrounding granulosa cells in primordial and primary follicles. Hoechst staining identifies the nucleus of cells. Scale bars; 30 μm.
Figure 5
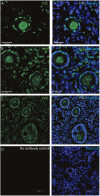
Intra-ovarian distribution of FUS in human primordial, and primary follicles. The FUS protein was detectable in (A) primordial follicles, (B,C) primary follicles (C) late pre-antral/early antral follicles. All samples were compared to a (D) no-antibody control. Hoechst staining identifies the nucleus of cells. Scale bars; 30 μm.
Similar articles
- Granulosa cells from human primordial and primary follicles show differential global gene expression profiles.
Ernst EH, Franks S, Hardy K, Villesen P, Lykke-Hartmann K. Ernst EH, et al. Hum Reprod. 2018 Apr 1;33(4):666-679. doi: 10.1093/humrep/dey011. Hum Reprod. 2018. PMID: 29506120 - Dormancy and activation of human oocytes from primordial and primary follicles: molecular clues to oocyte regulation.
Ernst EH, Grøndahl ML, Grund S, Hardy K, Heuck A, Sunde L, Franks S, Andersen CY, Villesen P, Lykke-Hartmann K. Ernst EH, et al. Hum Reprod. 2017 Aug 1;32(8):1684-1700. doi: 10.1093/humrep/dex238. Hum Reprod. 2017. PMID: 28854595 - Differentially Expressed lncRNAs After the Activation of Primordial Follicles in Mouse.
Zheng L, Luo R, Su T, Hu L, Gao F, Zhang X. Zheng L, et al. Reprod Sci. 2019 Aug;26(8):1094-1104. doi: 10.1177/1933719118805869. Epub 2018 Oct 30. Reprod Sci. 2019. PMID: 30376771 - Paraspeckle formation during the biogenesis of long non-coding RNAs.
Naganuma T, Hirose T. Naganuma T, et al. RNA Biol. 2013 Mar;10(3):456-61. doi: 10.4161/rna.23547. Epub 2013 Jan 16. RNA Biol. 2013. PMID: 23324609 Free PMC article. Review. - Biological Function of Long Non-coding RNA (LncRNA) Xist.
Wang W, Min L, Qiu X, Wu X, Liu C, Ma J, Zhang D, Zhu L. Wang W, et al. Front Cell Dev Biol. 2021 Jun 10;9:645647. doi: 10.3389/fcell.2021.645647. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34178980 Free PMC article. Review.
Cited by
- lncRNA FDNCR promotes apoptosis of granulosa cells by targeting the miR-543-3p/DCN/TGF-β signaling pathway in Hu sheep.
Yao X, Gao X, Bao Y, El-Samahy MA, Yang J, Wang Z, Li X, Zhang G, Zhang Y, Liu W, Wang F. Yao X, et al. Mol Ther Nucleic Acids. 2021 Mar 1;24:223-240. doi: 10.1016/j.omtn.2021.02.030. eCollection 2021 Jun 4. Mol Ther Nucleic Acids. 2021. PMID: 33767918 Free PMC article. - ITGB1-DT Facilitates Lung Adenocarcinoma Progression via Forming a Positive Feedback Loop With ITGB1/Wnt/β-Catenin/MYC.
Chang R, Xiao X, Fu Y, Zhang C, Zhu X, Gao Y. Chang R, et al. Front Cell Dev Biol. 2021 Mar 4;9:631259. doi: 10.3389/fcell.2021.631259. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33763420 Free PMC article. - Influence of Long Non-Coding RNAs on Human Oocyte Development.
Wang L, Li B, Cheng D. Wang L, et al. Pharmgenomics Pers Med. 2024 Jul 1;17:337-345. doi: 10.2147/PGPM.S449101. eCollection 2024. Pharmgenomics Pers Med. 2024. PMID: 38979513 Free PMC article. Review. - SPINT1-AS1 Drives Cervical Cancer Progression via Repressing miR-214 Biogenesis.
Song H, Liu Y, Liang H, Jin X, Liu L. Song H, et al. Front Cell Dev Biol. 2021 Jul 19;9:691140. doi: 10.3389/fcell.2021.691140. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34350182 Free PMC article. - NSUN5/TET2-directed chromatin-associated RNA modification of 5-methylcytosine to 5-hydroxymethylcytosine governs glioma immune evasion.
Wu R, Sun C, Chen X, Yang R, Luan Y, Zhao X, Yu P, Luo R, Hou Y, Tian R, Bian S, Li Y, Dong Y, Liu Q, Dai W, Fan Z, Yan R, Pan B, Feng S, Wu J, Chen F, Yang C, Wang H, Dai H, Shu M. Wu R, et al. Proc Natl Acad Sci U S A. 2024 Apr 2;121(14):e2321611121. doi: 10.1073/pnas.2321611121. Epub 2024 Mar 28. Proc Natl Acad Sci U S A. 2024. PMID: 38547058 Free PMC article.
References
- Andersson M. K., Ståhlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., et al. . (2008). The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 9:37. 10.1186/1471-2121-9-37 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources