Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells - PubMed (original) (raw)
Review
Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells
Samuel Wagner et al. FEMS Microbiol Lett. 2018.
Abstract
Virulence-associated type III secretion systems (T3SS) serve the injection of bacterial effector proteins into eukaryotic host cells. They are able to secrete a great diversity of substrate proteins in order to modulate host cell function, and have evolved to sense host cell contact and to inject their substrates through a translocon pore in the host cell membrane. T3SS substrates contain an N-terminal signal sequence and often a chaperone-binding domain for cognate T3SS chaperones. These signals guide the substrates to the machine where substrates are unfolded and handed over to the secretion channel formed by the transmembrane domains of the export apparatus components and by the needle filament. Secretion itself is driven by the proton motive force across the bacterial inner membrane. The needle filament measures 20-150 nm in length and is crowned by a needle tip that mediates host-cell sensing. Secretion through T3SS is a highly regulated process with early, intermediate and late substrates. A strict secretion hierarchy is required to build an injectisome capable of reaching, sensing and penetrating the host cell membrane, before host cell-acting effector proteins are deployed. Here, we review the recent progress on elucidating the assembly, structure and function of T3SS injectisomes.
Figures
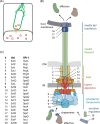
Figure 1.
Structure and function of the T3SS injectisome. A. Bacteria utilize T3SS to inject bacterial effector proteins into eukaryotic host cells. B. Structure of the T3SS indicating the different structural units (color-coded) and the individual components. Protein names are given in C. C. Nomenclature of T3SS components. The unified nomenclature (Uni) according to Hueck (1998) is shown on the left, the nomenclature of the T3SS encoded by Salmonella pathogenicity island 1 (SPI-1) is shown on the right. The nomenclature of the most commonly studied systems in Yersinia and pathogenic Escherichia coli follow the unified nomenclature with Ysc and Esc, respectively, instead of Sct. Abbreviations: IM, inner membrane; OM, outer membrane.
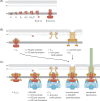
Figure 2.
Assembly of the T3SS injectisome. Protein names are indicated by their last letter, omitting ‘Sct’. The assembly pathways are shown from left to right. A. Assembly of the inner membrane export apparatus starts with membrane-integrated SctR. SctR (and also SctS and SctT, not shown) folds into a conformation that permits helical assembly. Then five SctR, followed by one SctT and four SctS, assemble into a helical assembly, winding up from the membrane. Recruitment of one SctU and then nine SctV finish the export apparatus assembly. B. Outside-in assembly model assuming independent secretin assembly. The export apparatus (the last step in A.) recruits 24 subunits of the inner inner ring protein SctJ. In an independent step, 15 secretin subunits assemble, supported by pilotins, into a pre-pore. PG penetration of the secretin is facilitated by hole-forming PG lytic enzymes. The secretin forms a pore in the outer membrane and recruits the outer inner ring protein SctD. Closure of the outer inner ring is not permitted until the SctJ-export apparatus assembly has been incorporated. C. Inside-out assembly model. The export apparatus (the last step in A.) recruits 24 SctJ and 24 SctD, followed by recruitment of the cytoplasmic components. Type III-dependent secretion of early substrates starts, which locally activates PG lytic enzymes in the periplasm to form a hole for secretin assembly. Just above the inner membrane complex, 15 secretin subunits assemble, supported by pilotins, into a pre-pore. Secretin pore formation and subsequent assembly of the needle filament complete the assembly of the injectisome. Abbreviations: IM, inner membrane; OM, outer membrane; PG, peptidoglycan.
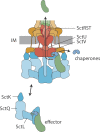
Figure 3.
The secretion pathway through the T3SS injectisome. Chaperone-substrate complexes are loaded onto free complexes of cytoplasmic components. These complexes are believed to serve as dynamic substrate carriers and deliver chaperone-substrate complexes to the injectisome. Here, chaperone-substrates complexes bind to the cytoplasmic domain of the major export apparatus protein SctV. Supported by the action of the ATPase, substrates are dechaperoned, unfolded and inserted into the secretion channel formed by the export apparatus components and the needle filament. Abbreviation: IM, inner membrane.
Similar articles
- Topological Analysis of the Type 3 Secretion System Translocon Pore Protein IpaC following Its Native Delivery to the Plasma Membrane during Infection.
Russo BC, Duncan JK, Goldberg MB. Russo BC, et al. mBio. 2019 May 28;10(3):e00877-19. doi: 10.1128/mBio.00877-19. mBio. 2019. PMID: 31138750 Free PMC article. - The Injectisome, a Complex Nanomachine for Protein Injection into Mammalian Cells.
Lara-Tejero M, Galán JE. Lara-Tejero M, et al. EcoSal Plus. 2019 Mar;8(2):10.1128/ecosalplus.ESP-0039-2018. doi: 10.1128/ecosalplus.ESP-0039-2018. EcoSal Plus. 2019. PMID: 30942149 Free PMC article. Review. - Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium.
Monjarás Feria JV, Lefebre MD, Stierhof YD, Galán JE, Wagner S. Monjarás Feria JV, et al. mBio. 2015 Oct 13;6(5):e01459-15. doi: 10.1128/mBio.01459-15. mBio. 2015. PMID: 26463164 Free PMC article. - The Tip Complex: From Host Cell Sensing to Translocon Formation.
Picking WD, Barta ML. Picking WD, et al. Curr Top Microbiol Immunol. 2020;427:173-199. doi: 10.1007/82_2019_171. Curr Top Microbiol Immunol. 2020. PMID: 31218507 Review. - Chaperone-mediated secretion switching from early to middle substrates in the type III secretion system encoded by Salmonella pathogenicity island 2.
Takaya A, Takeda H, Tashiro S, Kawashima H, Yamamoto T. Takaya A, et al. J Biol Chem. 2019 Mar 8;294(10):3783-3793. doi: 10.1074/jbc.RA118.005072. Epub 2019 Jan 16. J Biol Chem. 2019. PMID: 30651351 Free PMC article.
Cited by
- Cytosolic delivery of monobodies using the bacterial type III secretion system inhibits oncogenic BCR: ABL1 signaling.
Lebon C, Grossmann S, Mann G, Lindner F, Koide A, Koide S, Diepold A, Hantschel O. Lebon C, et al. Cell Commun Signal. 2024 Oct 16;22(1):500. doi: 10.1186/s12964-024-01874-6. Cell Commun Signal. 2024. PMID: 39415233 Free PMC article. - Physiochemical interaction between osmotic stress and a bacterial exometabolite promotes plant disease.
Getzke F, Wang L, Chesneau G, Böhringer N, Mesny F, Denissen N, Wesseler H, Adisa PT, Marner M, Schulze-Lefert P, Schäberle TF, Hacquard S. Getzke F, et al. Nat Commun. 2024 May 28;15(1):4438. doi: 10.1038/s41467-024-48517-5. Nat Commun. 2024. PMID: 38806462 Free PMC article. - Burkholderia pseudomallei Complex Subunit and Glycoconjugate Vaccines and Their Potential to Elicit Cross-Protection to Burkholderia cepacia Complex.
Badten AJ, Torres AG. Badten AJ, et al. Vaccines (Basel). 2024 Mar 15;12(3):313. doi: 10.3390/vaccines12030313. Vaccines (Basel). 2024. PMID: 38543947 Free PMC article. Review. - The sequence of events of enteropathogenic E. c oli's type III secretion system translocon assembly.
Gershberg J, Morhaim M, Rostrovsky I, Eichler J, Sal-Man N. Gershberg J, et al. iScience. 2024 Feb 3;27(3):109108. doi: 10.1016/j.isci.2024.109108. eCollection 2024 Mar 15. iScience. 2024. PMID: 38375228 Free PMC article. - Myxococcus xanthus predation: an updated overview.
Contreras-Moreno FJ, Pérez J, Muñoz-Dorado J, Moraleda-Muñoz A, Marcos-Torres FJ. Contreras-Moreno FJ, et al. Front Microbiol. 2024 Jan 24;15:1339696. doi: 10.3389/fmicb.2024.1339696. eCollection 2024. Front Microbiol. 2024. PMID: 38328431 Free PMC article. Review.
References
- Akeda Y, Galán JE. Chaperone release and unfolding of substrates in type III secretion. Nature 2005;437:911–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources