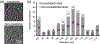Visualizing Individual RuBisCO and Its Assembly into Carboxysomes in Marine Cyanobacteria by Cryo-Electron Tomography - PubMed (original) (raw)
Visualizing Individual RuBisCO and Its Assembly into Carboxysomes in Marine Cyanobacteria by Cryo-Electron Tomography
Wei Dai et al. J Mol Biol. 2018.
Abstract
Cyanobacteria are photosynthetic organisms responsible for ~25% of the organic carbon fixation on earth. A key step in carbon fixation is catalyzed by ribulose bisphosphate carboxylase/oxygenase (RuBisCO), the most abundant enzyme in the biosphere. Applying Zernike phase-contrast electron cryo-tomography and automated annotation, we identified individual RuBisCO molecules and their assembly intermediates leading to the formation of carboxysomes inside Syn5 cyanophage infected cyanobacteria Synechococcus sp. WH8109 cells. Surprisingly, more RuBisCO molecules were found to be present as cytosolic free-standing complexes or clusters than as packaged assemblies inside carboxysomes. Cytosolic RuBisCO clusters and partially assembled carboxysomes identified in the cell tomograms support a concurrent assembly model involving both the protein shell and the enclosed RuBisCO. In mature carboxysomes, RuBisCO is neither randomly nor strictly icosahedrally packed within protein shells of variable sizes. A time-averaged molecular dynamics simulation showed a semi-liquid probability distribution of the RuBisCO in carboxysomes and correlated well with carboxysome subtomogram averages. Our structural observations reveal the various stages of RuBisCO assemblies, which could be important for understanding cellular function.
Keywords: RuBisCO; carboxysome biogenesis; convolutional neural network-based annotation; molecular dynamics simulation; zernike phase-contrast cryo-electron tomography.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures
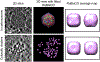
Figure 1.. Zernike phase contrast cryoET enables direct visualization of subcellular structures in Synechococcus sp. WH8109 cells.
(a) A 54Å slab taken from a WH8109 cell tomogram. (b) Volume rendering of the entire cell tomogram shown in (a). Carboxysome shell is segmented and colored in orange. RuBisCO complexes are colored in magenta. (c) Zoom-in view of the region boxed in (a).
Figure 2.. Applying automated neural-network based annotation and subtomogram analysis to study RuBisCO distribution and packing in individual carboxysomes.
Left: slice views of a carboxysome and a cytosolic RuBisCO cluster; Middle: isosurface views of a carboxysome and of a RuBisCO cluster. The carboxysome is radially colored with blue representing the innermost layer of RuBisCO, green, representing the middle layer, and red to represent the outermost layer. Right: top and side views of RuBisCO subtomogram averages from carboxysomes (top) and from cytosolic clusters (bottom).
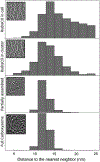
Figure 3.. Comparison of distance to the nearest neighbors of RuBisCO complexes inside WH8109 cells.
The insets are representative clusters or particles in each class.
Figure 4.. Carboxysome size distribution and the numbers of icosahedral and non-icosahedral particles in each size group.
(a) Central slabs of 54 Å of representative icosahedral and non-icosahedral particles. (b) Carboxysome size distribution histogram including both non-symmetric and icosahedral particles in each size group. Intracellular carboxysome subtomograms were classified into various size groups based on their diameters by radial density plotting.
Figure 5.. Partially assembled carboxysomes identified in Synechococcus WH8109 cell tomograms.
(a) Section view of a tomogram of a cell that has just completed cell division showing a partially assembled carboxysome labeled by “C”. (b) Slice and color isosurface views of partially assembled carboxysomes. The carboxysomes were arranged from particles with a small number of RuBisCO and shell units, probably representing particles in the early stage of carboxysome biogenesis, to particles that are close to completion of assembly. Magenta: shell; yellow: RuBisCO.
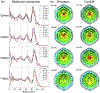
Figure 6.. Molecular dynamic simulation vs experimental subtomogram averages of carboxysomes.
(a) Radial density plots of individual carboxysome simulations with variable numbers of RuBisCO (N) enclosed were compared to those of experimental subtomogram averages (red line). The red dashed lines denote the locations of the density peaks in the radial density plots. The cross sectional views of best matching simulation maps were shown with corresponding subtomogram average maps in (b).
Similar articles
- Rubisco accumulation factor 1 (Raf1) plays essential roles in mediating Rubisco assembly and carboxysome biogenesis.
Huang F, Kong WW, Sun Y, Chen T, Dykes GF, Jiang YL, Liu LN. Huang F, et al. Proc Natl Acad Sci U S A. 2020 Jul 21;117(29):17418-17428. doi: 10.1073/pnas.2007990117. Epub 2020 Jul 7. Proc Natl Acad Sci U S A. 2020. PMID: 32636267 Free PMC article. - Structure and assembly of cargo Rubisco in two native α-carboxysomes.
Ni T, Sun Y, Burn W, Al-Hazeem MMJ, Zhu Y, Yu X, Liu LN, Zhang P. Ni T, et al. Nat Commun. 2022 Jul 25;13(1):4299. doi: 10.1038/s41467-022-32004-w. Nat Commun. 2022. PMID: 35879301 Free PMC article. - Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA.
Long BM, Badger MR, Whitney SM, Price GD. Long BM, et al. J Biol Chem. 2007 Oct 5;282(40):29323-35. doi: 10.1074/jbc.M703896200. Epub 2007 Aug 3. J Biol Chem. 2007. PMID: 17675289 - Cyanobacterial carboxysomes: microcompartments that facilitate CO2 fixation.
Rae BD, Long BM, Whitehead LF, Förster B, Badger MR, Price GD. Rae BD, et al. J Mol Microbiol Biotechnol. 2013;23(4-5):300-7. doi: 10.1159/000351342. Epub 2013 Aug 5. J Mol Microbiol Biotechnol. 2013. PMID: 23920493 Review. - Atypical Carboxysome Loci: JEEPs or Junk?
USF Genomics Class 2020; USF Genomics Class 2021; Sutter M, Kerfeld CA, Scott KM. USF Genomics Class 2020, et al. Front Microbiol. 2022 May 20;13:872708. doi: 10.3389/fmicb.2022.872708. eCollection 2022. Front Microbiol. 2022. PMID: 35668770 Free PMC article. Review.
Cited by
- Exploring Transient States of PAmKate to Enable Improved Cryogenic Single-Molecule Imaging.
Perez D, Dowlatshahi DP, Azaldegui CA, Ansell TB, Dahlberg PD, Moerner WE. Perez D, et al. J Am Chem Soc. 2024 Oct 23;146(42):28707-28716. doi: 10.1021/jacs.4c05632. Epub 2024 Oct 10. J Am Chem Soc. 2024. PMID: 39388715 Free PMC article. - Nanoengineering Carboxysome Shells for Protein Cages with Programmable Cargo Targeting.
Li T, Chang P, Chen W, Shi Z, Xue C, Dykes GF, Huang F, Wang Q, Liu LN. Li T, et al. ACS Nano. 2024 Mar 12;18(10):7473-7484. doi: 10.1021/acsnano.3c11559. Epub 2024 Feb 7. ACS Nano. 2024. PMID: 38326220 Free PMC article. - Uncovering the roles of the scaffolding protein CsoS2 in mediating the assembly and shape of the α-carboxysome shell.
Li T, Chen T, Chang P, Ge X, Chriscoli V, Dykes GF, Wang Q, Liu L-N. Li T, et al. mBio. 2024 Oct 16;15(10):e0135824. doi: 10.1128/mbio.01358-24. Epub 2024 Aug 29. mBio. 2024. PMID: 39207096 Free PMC article. - Extracellular CahB1 from Sodalinema gerasimenkoae IPPAS B-353 Acts as a Functional Carboxysomal β-Carbonic Anhydrase in Synechocystis sp. PCC6803.
Minagawa J, Dann M. Minagawa J, et al. Plants (Basel). 2023 Jan 6;12(2):265. doi: 10.3390/plants12020265. Plants (Basel). 2023. PMID: 36678979 Free PMC article. - Singular adaptations in the carbon assimilation mechanism of the polyextremophile cyanobacterium Chroococcidiopsis thermalis.
Aguiló-Nicolau P, Galmés J, Fais G, Capó-Bauçà S, Cao G, Iñiguez C. Aguiló-Nicolau P, et al. Photosynth Res. 2023 May;156(2):231-245. doi: 10.1007/s11120-023-01008-y. Epub 2023 Mar 20. Photosynth Res. 2023. PMID: 36941458 Free PMC article.
References
- Grotzschel S, de Beer D. Effect of oxygen concentration on photosynthesis and respiration in two hypersaline microbial mats. Microb Ecol. 2002;44:208–16. - PubMed
- Hackenberg C, Huege J, Engelhardt A, Wittink F, Laue M, Matthijs HC, et al. Low-carbon acclimation in carboxysome-less and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology. 2012;158:398–413. - PubMed
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, et al. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–6. - PubMed
- Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. Cyanobacterial-based approaches to improving photosynthesis in plants. J of Exp Bot. 2013;64:787–98. - PubMed
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–22. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources