The germline genetic component of drug sensitivity in cancer cell lines - PubMed (original) (raw)
The germline genetic component of drug sensitivity in cancer cell lines
Michael P Menden et al. Nat Commun. 2018.
Abstract
Patients with seemingly the same tumour can respond very differently to treatment. There are strong, well-established effects of somatic mutations on drug efficacy, but there is at-most anecdotal evidence of a germline component to drug response. Here, we report a systematic survey of how inherited germline variants affect drug susceptibility in cancer cell lines. We develop a joint analysis approach that leverages both germline and somatic variants, before applying it to screening data from 993 cell lines and 265 drugs. Surprisingly, we find that the germline contribution to variation in drug susceptibility can be as large or larger than effects due to somatic mutations. Several of the associations identified have a direct relationship to the drug target. Finally, using 17-AAG response as an example, we show how germline effects in combination with transcriptomic data can be leveraged for improved patient stratification and to identify new markers for drug sensitivity.
Conflict of interest statement
U.M. is a founder, stockholder and consultant for 14M Genomics Ltd. M.P.M. and U.M. are employees of AstraZeneca. The remaining authors declare no competing interests.
Figures
Fig. 1
Illustration of the joint analysis approach considering germline variants and somatic mutations. a Prediction of drug susceptibility, either exclusively considering somatic mutations (baseline, black line) or considering the combination of germline variants and somatic mutations (green). Shown is out-of-sample prediction performance measured by the Pearson correlation coefficient between predicted and observed drug susceptibility profiles (quantified as 1-AUC; Methods). Error bars show standard deviations across analysis repetitions of the difference of Pearson correlation coefficients from the compared models (Methods, Supplementary Note 1). Selected drugs with large improvements of prediction performance when accounting for germline variants are highlighted. b Illustration of a joint genome-wide association analysis, considering associations between somatic mutations (green) or germline variants (black) and drug susceptibility for 17-AAG. Germline variants with genome-wide significant associations are highlighted in red (FWER < 0.05, dashed horizontal line)
Fig. 2
Germline and somatic associations with drug susceptibility. a Negative log P values from genome-wide association analyses with drug susceptibility phenotypes of 265 drugs, either considering lead germline variants (_x_-axis) or lead somatic mutations (_y_-axis). Shown are lead associations, i.e., the most significant association for either QTL type for each drug. b Effect size estimates for the associations shown in a, considering the corresponding lead germline associations (_x_-axis) or lead somatic associations (_y_-axis). Each dot represents a drug. Drugs coloured in blue have a significant germline or somatic association (FWER < 0.05). Somatic QTLs tended to have large effect sizes than germline QTLs. (See also Supplementary Fig. 5 for an analysis stratified by variant frequency)
Fig. 3
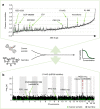
The germline component of 17-AAG drug susceptibility. a Quantile–quantile plot of negative log P values from genome-wide association tests of 17-AAG susceptibility, considering germline variants (back) and somatic mutations (green). The most associated (lead) variant rs12595927 is in tight linkage disequilibrium with the known causal variant for this associations (rs1800566, e.g., ref., _r_2 = 1 in European populations). b Scatter plot between NQO1 gene expression (_x_-axis) and 17-AAG drug susceptibility (_y_-axis). Dots correspond to individual cell lines stratified by genotype at the rs1800566 locus (yellow: TT allele, blue: CC/CT allele). Box plots show the effect of the rs1800566 locus on gene expression (top panel) and drug susceptibility (right panel). Whereas NQO1 expression level is not associated with the germline variant, rs1800566 modulates the association between NQO1 expression level and drug susceptibility. The combination of high expression levels of NQO1 together with a CC or CT genotype is associated with the largest drug response. Boxes extend from the lower quartile (Q1) of the data to the upper quartile (Q2) of the data, whiskers show the range of the data (after excluding outliers), fliers show outliers and the red lines show the medians. Outliers are defined by the standard condition x < Q1 −1.5(Q2 −Q1) ∨ _x_ > Q2 + 1.5(Q2 −Q1). c Scatter plot between NQO1 expression level (_x_-axis) and 17-AAG drug susceptibility (_y_-axis), stratified by tissue type. d Lower panel: Mean drug efficacy of AUY922 and 17-AAG as a function of NQO1 expression and stratified for rs1800566 genotype. Colours indicated the genotype group with triangles corresponding to AUY922 response and circles denoting 17-AAG response. Shaded areas indicate plus or minus one standard error of the mean drug response. Top panel: number of cell lines in stratified groups. AUY922 is more effective than 17-AAG for low NQO1 expression. In cell lines with a CT and CC (rs1800566) germline background and high expression of NQO1, the efficacy of 17-AAG is comparable or larger than AUY922. e Mode of action for 17-AAG and AUY922. f Frequency of NQO1 germline variants in different human populations. Data for rs1800566 extracted from EnsEMBL
Similar articles
- A method to reduce ancestry related germline false positives in tumor only somatic variant calling.
Halperin RF, Carpten JD, Manojlovic Z, Aldrich J, Keats J, Byron S, Liang WS, Russell M, Enriquez D, Claasen A, Cherni I, Awuah B, Oppong J, Wicha MS, Newman LA, Jaigge E, Kim S, Craig DW. Halperin RF, et al. BMC Med Genomics. 2017 Oct 19;10(1):61. doi: 10.1186/s12920-017-0296-8. BMC Med Genomics. 2017. PMID: 29052513 Free PMC article. - A New Frontier for Cancer Genetics: Identification of Germline-Somatic Associations.
Kar SP. Kar SP. Cancer Res. 2023 Apr 14;83(8):1165-1166. doi: 10.1158/0008-5472.CAN-23-0152. Cancer Res. 2023. PMID: 37057597 - Small molecule inhibitor screening identifified HSP90 inhibitor 17-AAG as potential therapeutic agent for gallbladder cancer.
Weber H, Valbuena JR, Barbhuiya MA, Stein S, Kunkel H, García P, Bizama C, Riquelme I, Espinoza JA, Kurtz SE, Tyner JW, Calderon JF, Corvalán AH, Grez M, Pandey A, Leal-Rojas P, Roa JC. Weber H, et al. Oncotarget. 2017 Apr 18;8(16):26169-26184. doi: 10.18632/oncotarget.15410. Oncotarget. 2017. PMID: 28412732 Free PMC article. - The Evolutionary Interplay of Somatic and Germline Mutation Rates.
Beichman AC, Zhu L, Harris K. Beichman AC, et al. Annu Rev Biomed Data Sci. 2024 Aug;7(1):83-105. doi: 10.1146/annurev-biodatasci-102523-104225. Epub 2024 Jul 24. Annu Rev Biomed Data Sci. 2024. PMID: 38669515 Review. - Germline genetic variation, cancer outcome, and pharmacogenetics.
Coate L, Cuffe S, Horgan A, Hung RJ, Christiani D, Liu G. Coate L, et al. J Clin Oncol. 2010 Sep 10;28(26):4029-37. doi: 10.1200/JCO.2009.27.2336. Epub 2010 Aug 2. J Clin Oncol. 2010. PMID: 20679599 Review.
Cited by
- Advancing precision oncology with AI-powered genomic analysis.
Srivastava R. Srivastava R. Front Pharmacol. 2025 Apr 30;16:1591696. doi: 10.3389/fphar.2025.1591696. eCollection 2025. Front Pharmacol. 2025. PMID: 40371349 Free PMC article. Review. - Individualized Prediction of Drug Response and Rational Combination Therapy in NSCLC Using Artificial Intelligence-Enabled Studies of Acute Phosphoproteomic Changes.
Coker EA, Stewart A, Ozer B, Minchom A, Pickard L, Ruddle R, Carreira S, Popat S, O'Brien M, Raynaud F, de Bono J, Al-Lazikani B, Banerji U. Coker EA, et al. Mol Cancer Ther. 2022 Jun 1;21(6):1020-1029. doi: 10.1158/1535-7163.MCT-21-0442. Mol Cancer Ther. 2022. PMID: 35368084 Free PMC article. - Bioinformatics roadmap for therapy selection in cancer genomics.
Jiménez-Santos MJ, García-Martín S, Fustero-Torre C, Di Domenico T, Gómez-López G, Al-Shahrour F. Jiménez-Santos MJ, et al. Mol Oncol. 2022 Nov;16(21):3881-3908. doi: 10.1002/1878-0261.13286. Epub 2022 Aug 20. Mol Oncol. 2022. PMID: 35811332 Free PMC article. Review. - MKX-AS1 Gene Expression Associated with Variation in Drug Response to Oxaliplatin and Clinical Outcomes in Colorectal Cancer Patients.
Gonzalez RD, Small GW, Green AJ, Akhtari FS, Motsinger-Reif AA, Quintanilha JCF, Havener TM, Reif DM, McLeod HL, Wiltshire T. Gonzalez RD, et al. Pharmaceuticals (Basel). 2023 May 17;16(5):757. doi: 10.3390/ph16050757. Pharmaceuticals (Basel). 2023. PMID: 37242540 Free PMC article. - RYK Gene Expression Associated with Drug Response Variation of Temozolomide and Clinical Outcomes in Glioma Patients.
Gonzalez RD, Small GW, Green AJ, Akhtari FS, Havener TM, Quintanilha JCF, Cipriani AB, Reif DM, McLeod HL, Motsinger-Reif AA, Wiltshire T. Gonzalez RD, et al. Pharmaceuticals (Basel). 2023 May 10;16(5):726. doi: 10.3390/ph16050726. Pharmaceuticals (Basel). 2023. PMID: 37242509 Free PMC article.
References
- Seashore-Ludlow B, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5:1210–1223. doi: 10.1158/2159-8290.CD-15-0235. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources