Stereotypic Immune System Development in Newborn Children - PubMed (original) (raw)
Stereotypic Immune System Development in Newborn Children
Axel Olin et al. Cell. 2018.
Abstract
Epidemiological data suggest that early life exposures are key determinants of immune-mediated disease later in life. Young children are also particularly susceptible to infections, warranting more analyses of immune system development early in life. Such analyses mostly have been performed in mouse models or human cord blood samples, but these cannot account for the complex environmental exposures influencing human newborns after birth. Here, we performed longitudinal analyses in 100 newborn children, sampled up to 4 times during their first 3 months of life. From 100 μL of blood, we analyze the development of 58 immune cell populations by mass cytometry and 267 plasma proteins by immunoassays, uncovering drastic changes not predictable from cord blood measurements but following a stereotypic pattern. Preterm and term children differ at birth but converge onto a shared trajectory, seemingly driven by microbial interactions and hampered by early gut bacterial dysbiosis.
Keywords: CyTOF; human immunology; immune system development; immune variation; mass cytometry; neonate; neonatology; newborn immune systems; preterm birth; systems immunology.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
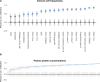
Preterm and Term Children Differ at Birth (A) Blood samples of 100 newborn children and their parents were collected at up to four different time points during the first months of life. Plasma protein concentrations were measured by ProSeek, and immune cells were analyzed by mass cytometry. (B) Median protein concentrations in cord blood of term and preterm children. Differently regulated proteins (false discovery rate [FDR] < 0.01) are marked in blue. (C) Intersample differences between cord blood samples from preterm and term children were visualized by multidimensional scaling (MDS). (D) Gestational age at birth for preterm and term children and preterm children at 3 months postnatal age. (E and F) Interindividual distances between term and preterm children at birth (gray) or term children at birth and preterm children at 3 months after birth (blue). Interindividual distances separately calculated for plasma proteins (E) (Euclidian distance) and cell composition (F) (Aitchison’s distance). Error bars represent ±1.5 IGR above and below Q1 and Q3, respectively. See also Figure S1 and Tables S2, S3, and S4.
Figure S1
Cord Blood Immune Parameters in Preterm and Term Children, Related to Figure 1 (A) Principle component analysis of plasma protein concentrations in cord blood of term (blue) and preterm (pink) children. (B) Neutrophil levels as a fraction of all white blood cells in cord blood in relation to gestational age at birth. A linear regression curve is fit to the data (blue line) together with a confidence interval (light blue shade).
Figure 2
Cord Blood Is Not Representative of Postnatal Immunity (A) Pearson’s correlation coefficients of immune cell frequencies in cord blood versus 1-week blood (gray), nonsense control correlation between random samples (black), and significantly different features (orange). Error bars represent confidence intervals. (B) Intersample differences between cord blood samples, as well as peripheral blood samples collected on the day of birth and the following days, visualized by MDS. Sampling day is indicated. CB, cord blood. MDS coordinates are based on pairwise Aitchison’s distances (cell composition). (C) Pearson’s correlation coefficients of plasma protein concentrations in cord blood versus 1-week blood (gray nonsense control correlation between random samples (black) and significantly different features (orange). Error bars represent confidence intervals. (D) bhSNE embedding of indicated cells in blood samples from two consecutive weeks in an adult. (E) bhSNE maps of immune cell phenotypes in cord blood and week 1 from a newborn. (F) Pairwise Jensen-Shannon (JS) distances between bhSNE embeddings for all time points from adult samples (gray) and for cord blood versus 1-week newborn samples (orange). p values for comparisons of the mean distances between groups. See also Figure S2.
Figure S2
Over Time Stability of Immune Parameters in Healthy Adults, Related to Figure 2 (A and B) Pearson’s correlation coefficients of immune cell frequencies (A) and plasma protein concentrations (B) between two peripheral blood samples taken 3 months apart in a cohort of 100 healthy individuals. Included is also a nonsense control correlation between random samples (black). Cell populations (A) or plasma protein concentrations (B) that are significantly correlated compared to control samples are indicated in blue.
Figure 3
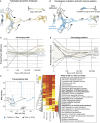
Topological Data Analysis Reveals Systems-Level Convergence of Term and Preterm Immune Systems during the First Weeks of Life (A) A parameter landscape model by topological data analysis (TDA) using frequencies of 48 cell populations and 250 plasma protein concentrations from 202 samples of newborn children (37 cord blood, 68 week-1, 38 week-4, and 59 week-12 samples). Each node in the network represents a set of correlated samples; each sample can be included in multiple nodes; and nodes sharing at least one sample connected with an edge. The network is colored by the average sampling day of its samples. (B) Nodes colored by the proportion of samples from term and preterm children. (C and D) Differences in immune cell frequencies (C) and plasma protein concentrations (D) between preterm and term children separated into different age groups. (E) Principal component analysis of mRNA-seq transcripts per kilobase million (TPM) values from preterm (n = 4) and term (n = 4) children sampled at weeks 1 and 12 and two adult control samples. (F) Differential expression analysis of RNA-seq data between preterm and term children at 12 weeks. GO terms are listed for genes significantly upregulated in preterm versus term children. See also Figure S3.
Figure S3
Global Immune System Development in Relation to Metadata, Related to Figure 3 (A) TDA network landscape from Figure 3A showing the number of samples included in each node of the network. (B) The same network as in (A) and colored by the proportion of samples in the node taken from children delivered by cesarean section or vaginal delivery respectively. (C) PCA analysis of plasma protein showing PC1 (x axis) and PC2 (y axis) and individual samples colored by sex. (D) MDS analysis of cell composition showing component 1 (x axis) and 2 (y axis) and individual samples colored by sex. (E) PCA analysis of plasma protein showing PC1 (x axis) and PC2 (y axis) and individual samples colored by season of sampling. (F) MDS analysis of cell composition showing component 1 (x axis) and 2 (y axis) and individual samples colored by season of sampling.
Figure 4
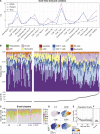
A Patterned Progression in Immune Cell Composition (A) The coefficient of variance (CV) for 21 gated cell populations in healthy adults (n = 3), preterm (n = 29), and term (n = 15) children. (B) The relative proportions of 10 cell populations in 183 postnatal blood samples from 57 children ordered by sampling day of life. (C) Composition of immune cell clusters analyzed using self-organizing map clustering of the B cell population. (D) The B cell cluster #6 on a bhSNE analysis of total B cells and relative CD38, CD9, and CD24 expression. (E) The relative frequency of cluster B #6 over time in children. Shaded area represents the confidence interval around the mean. (F) Frequency of cluster B #6 in parents. See also Figure S4.
Figure S4
Immune Cell Population Variance Early in Life, Related to Figure 4 (A and B) The relative contribution to the total variance of inter- and intra-individual variance for 24 immune cell populations in newborn children (A) and healthy adults (B). (C–G) Composition of immune cell clusters analyzed separately using self-organizing map clustering, within CD4+ T cells (C), CD8+ T cells (D), NK-cells (E), neutrophils (F), and monocytes (G). (H) The Monocyte cluster #8 is highlighted in a bhSNE map within total CD14+ monocytes and the relative expression of CD14, HLA-DR and CD31 is highlighted. (I and J) The relative frequency of cluster M #8 over time in children (I), and (J) parents.
Figure 5
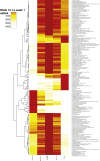
Phenotypic Development of Immune Cell Populations (A) TDA network landscape with maternal and newborn samples combined. (B) bhSNE-maps of five cell populations from one child (orange) and its parents (gray). (C) JS distances between phenotypes in each child and its parents separately calculated and plotted against gestational age (days) at the time of sampling. Linear regression curves with confidence intervals are shown, and R2 values and p values for each correlation. (D) Boxplots of the JS distances for cell phenotypes between coparents. Error bars represent ±1.5 IGR above and below Q1 and Q3, respectively.
Figure 6
Immune Parameter Changes during the First Weeks of Life (A) The TDA parameter landscape subdivided into five developmental stages. (B and C) Interindividual distances for plasma protein profiles (B) (Euclidean) and cell composition (C) (Atchinson’s) across the five stages. (D) Plasma concentration (log2 NPX) of cytokines IL-27 and IL-10. (E) Plasma concentration (log2 NPX) of PIgR. (F) Plasma concentration (log2 NPX) of IL-8 (CXCL8) and cytokines IL-17A and IL-12B. (G) The frequency of CD8+ T cell cluster #1 as a fraction of all cells. (H) bhSNE plots showing cluster localization among CD8+ T cells and CD3, CD8a, CD161, and CD38 expression. All p values from KS tests comparing distributions of stages 2–3 combined versus stage 5. (I) Differentially regulated genes week 12 versus week 1 associated with the GO: positive reg. of immune system process. Genes ordered by absolute log2 (week 12/week 1) (x axis) and actual log2 (week 12/week 1) (y axis). See also Figure S5 and Table S1.
Figure S5
Differentially Regulated Transcriptional Programs between Weeks 1 and 12 after Birth, Related to Figure 6 Gene ontology terms in week 12 versus week 1 from RNaseq-data between. GO-terms for genes significantly different week 12 versus week 1 are shown and divided by gene set overall direction of differential regulation; up only, down only, mixed, mostly up, mixed mostly down and unidirectional.
Figure 7
Immune System Development and Gut Bacterial composition (A) Principle coordinate analysis of bacterial composition in fecal samples (n = 95) collected at 1, 4, and 12 weeks of age in newborn children (n = 45). (B) Shannon α diversity in each sample and age at sampling. Individuals are divided into normal (purple, Shannon α > 0.3) and dysbiotic cases (orange, Shannon α < 0.3). (C) Bacterial class abundances in week 1 samples with normal and dysbiotic gut microbiome. (D) Bacterial class composition across fecal samples grouped by week of life and ordered within groups by PCoA2. Shaded area represents confidence interval around the mean. (E) Cell frequencies as log2(dysbiosis/normal) at 12 weeks, ranked by log2ratio. Top four highlighted and named. (F) Plasma protein concentrations as log2ratio (dysbiosis/normal) at 12 weeks, ranked by log2ratio. Top five highlighted and named. (G) Pairwise interindividual Aitchison’s distances between immune cell compositions at 12 weeks within the dysbiosis group (orange) and normal group (purple). Error bars represent ±1.5 IGR above and below Q1 and Q3, respectively. (H) One hypothetical adaptive change within state space induced by a single input. (I) Adaptation to the same stimuli is constrained when partially opposed by a second simultaneous input. (J) Convergence of diverse immune systems in state space by a large number of simultaneous inputs inducing opposing adaptive responses.
Comment in
- Neonate-omics: Charting the Unknown Immune Response in Early Life.
Jennewein MF, Butler AL, Alter G. Jennewein MF, et al. Cell. 2018 Aug 23;174(5):1051-1053. doi: 10.1016/j.cell.2018.08.001. Cell. 2018. PMID: 30142343 - Baby steps for the immune system.
Bordon Y. Bordon Y. Nat Rev Immunol. 2018 Oct;18(10):600-601. doi: 10.1038/s41577-018-0062-y. Nat Rev Immunol. 2018. PMID: 30166618 No abstract available. - Early immune development.
Stower H. Stower H. Nat Med. 2018 Oct;24(10):1491. doi: 10.1038/s41591-018-0226-0. Nat Med. 2018. PMID: 30297890 No abstract available.
Similar articles
- Immune tolerance attenuates gut dysbiosis, dysregulated uterine gene expression and high-fat diet potentiated preterm birth in mice.
Manuel CR, Latuga MS, Ashby CR Jr, Reznik SE. Manuel CR, et al. Am J Obstet Gynecol. 2019 Jun;220(6):596.e1-596.e28. doi: 10.1016/j.ajog.2019.02.028. Epub 2019 Feb 18. Am J Obstet Gynecol. 2019. PMID: 30790568 - Perinatal Microbiomes' Influence on Preterm Birth and Preterms' Health: Influencing Factors and Modulation Strategies.
Ruiz L, Moles L, Gueimonde M, Rodriguez JM. Ruiz L, et al. J Pediatr Gastroenterol Nutr. 2016 Dec;63(6):e193-e203. doi: 10.1097/MPG.0000000000001196. J Pediatr Gastroenterol Nutr. 2016. PMID: 27019409 Review. - Implementing Mass Cytometry at the Bedside to Study the Immunological Basis of Human Diseases: Distinctive Immune Features in Patients with a History of Term or Preterm Birth.
Gaudillière B, Ganio EA, Tingle M, Lancero HL, Fragiadakis GK, Baca QJ, Aghaeepour N, Wong RJ, Quaintance C, El-Sayed YY, Shaw GM, Lewis DB, Stevenson DK, Nolan GP, Angst MS. Gaudillière B, et al. Cytometry A. 2015 Sep;87(9):817-29. doi: 10.1002/cyto.a.22720. Epub 2015 Jul 17. Cytometry A. 2015. PMID: 26190063 Free PMC article. - Meta-Analysis of Maternal and Fetal Transcriptomic Data Elucidates the Role of Adaptive and Innate Immunity in Preterm Birth.
Vora B, Wang A, Kosti I, Huang H, Paranjpe I, Woodruff TJ, MacKenzie T, Sirota M. Vora B, et al. Front Immunol. 2018 May 9;9:993. doi: 10.3389/fimmu.2018.00993. eCollection 2018. Front Immunol. 2018. PMID: 29867970 Free PMC article. - Early nutrition and immunity - progress and perspectives.
Calder PC, Krauss-Etschmann S, de Jong EC, Dupont C, Frick JS, Frokiaer H, Heinrich J, Garn H, Koletzko S, Lack G, Mattelio G, Renz H, Sangild PT, Schrezenmeir J, Stulnig TM, Thymann T, Wold AE, Koletzko B. Calder PC, et al. Br J Nutr. 2006 Oct;96(4):774-90. Br J Nutr. 2006. PMID: 17010239
Cited by
- Mechanisms of microbe-mediated immune development in the context of antibiotics and asthma.
Donald K, Finlay BB. Donald K, et al. Front Allergy. 2024 Oct 14;5:1469426. doi: 10.3389/falgy.2024.1469426. eCollection 2024. Front Allergy. 2024. PMID: 39469482 Free PMC article. Review. - Perinatal development of innate immune topology.
Henneke P, Kierdorf K, Hall LJ, Sperandio M, Hornef M. Henneke P, et al. Elife. 2021 May 25;10:e67793. doi: 10.7554/eLife.67793. Elife. 2021. PMID: 34032570 Free PMC article. Review. - Analysis of microbial composition and sharing in low-biomass human milk samples: a comparison of DNA isolation and sequencing techniques.
Spreckels JE, Fernández-Pato A, Kruk M, Kurilshikov A, Garmaeva S, Sinha T, Ghosh H, Harmsen H, Fu J, Gacesa R, Zhernakova A. Spreckels JE, et al. ISME Commun. 2023 Nov 9;3(1):116. doi: 10.1038/s43705-023-00325-6. ISME Commun. 2023. PMID: 37945978 Free PMC article. - A decade of neonatal sepsis caused by gram-negative bacilli-a retrospective matched cohort study.
Nordberg V, Iversen A, Tidell A, Ininbergs K, Giske CG, Navér L. Nordberg V, et al. Eur J Clin Microbiol Infect Dis. 2021 Sep;40(9):1803-1813. doi: 10.1007/s10096-021-04211-8. Epub 2021 Mar 24. Eur J Clin Microbiol Infect Dis. 2021. PMID: 33761020 Free PMC article. - Prospective multicentre study of host response signatures in neonatal sepsis in Sub Saharan Africa.
Ezinmegnon S, Mommert M, Bartolo F, Agbota G, Darius S, Briand V, d'Almeida M, Alao MJ, Dossou-Dagba I, Massougbodji A, Lausten-Thomsen U, Pachot A, Vachot L, Yugueros-Marcos J, Brengel-Pesce K, Fievet N, Tissieres P. Ezinmegnon S, et al. Sci Rep. 2022 Dec 12;12(1):21458. doi: 10.1038/s41598-022-25892-x. Sci Rep. 2022. PMID: 36509812 Free PMC article.
References
- Alcaïs A., Quintana-Murci L., Thaler D.S., Schurr E., Abel L., Casanova J.L. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann. N Y Acad. Sci. 2010;1214:18–33. - PubMed
- Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., Kuzeljevic B., Gold M.J., Britton H.M., Lefebvre D.L., CHILD Study Investigators Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical