Partial genomic survival of cave bears in living brown bears - PubMed (original) (raw)
. 2018 Oct;2(10):1563-1570.
doi: 10.1038/s41559-018-0654-8. Epub 2018 Aug 27.
James A Cahill 2, Stefanie Hartmann 3, Christoph Theunert 4 5, Georgios Xenikoudakis 3, Gloria G Fortes 3 6, Johanna L A Paijmans 3, Gernot Rabeder 7, Christine Frischauf 7, Aurora Grandal-d'Anglade 8, Ana García-Vázquez 8, Marine Murtskhvaladze 9, Urmas Saarma 10, Peeter Anijalg 10, Tomaž Skrbinšek 11, Giorgio Bertorelle 6, Boris Gasparian 12, Guy Bar-Oz 13, Ron Pinhasi 14 15, Montgomery Slatkin 4, Love Dalén 16, Beth Shapiro 2, Michael Hofreiter 3
Affiliations
- PMID: 30150744
- PMCID: PMC6590514
- DOI: 10.1038/s41559-018-0654-8
Partial genomic survival of cave bears in living brown bears
Axel Barlow et al. Nat Ecol Evol. 2018 Oct.
Abstract
Although many large mammal species went extinct at the end of the Pleistocene epoch, their DNA may persist due to past episodes of interspecies admixture. However, direct empirical evidence of the persistence of ancient alleles remains scarce. Here, we present multifold coverage genomic data from four Late Pleistocene cave bears (Ursus spelaeus complex) and show that cave bears hybridized with brown bears (Ursus arctos) during the Pleistocene. We develop an approach to assess both the directionality and relative timing of gene flow. We find that segments of cave bear DNA still persist in the genomes of living brown bears, with cave bears contributing 0.9 to 2.4% of the genomes of all brown bears investigated. Our results show that even though extinction is typically considered as absolute, following admixture, fragments of the gene pool of extinct species can survive for tens of thousands of years in the genomes of extant recipient species.
Figures
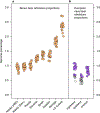
Fig. 1 |. Phylogenetic relationships of the sequenced brown, polar and cave bear genomes.
a, The maximum-likelihood phylogeny of individuals used in this study based on whole-genome transversion differences, rooted using the American black bear outgroup. b, Measures of clade differentiation based on D statistic tests inconsistent with the species tree. The tested topologies are shown on the left, with points showing the calculated clade differentiation (D values) for all possible combinations of the sampled individuals. This measure of clade differentiation scales between 0 (indicating a phylogenetic trifurcation) and 1 (indicating complete lineage sorting and absence of post-divergence gene flow). Measures of differentiation for the brown bear clade relative to polar bears, and for the polar bear clade relative to brown bears, are shown for comparison with values obtained for cave bears.
Fig. 2 |
a, The proportion of the genomes of individual brown bears derived from admixture with cave bears, following the divergence of brown bears and polar bears (_f̂_results). Eighteen admixture estimates per brown bear represent all possible combinations of European cave bear introgressor, relative to three polar bears. Almost all values are strongly significant (Z > 3). A small number of comparisons involving the Russian and American bears were more moderately supported (Z > 2.4). ABC, Admiralty, Baranof and Chichagof Islands; Den., Denali; LP, Late Pleistocene. b, The proportion of the genomes of individual European cave bears derived from admixture with brown bears, following the divergence of the European cave bear clade and the Caucasus cave bear kudarensis (_f̂_results). Twenty dots per cave bear sample represent all possible combinations of modern Eurasian brown bear introgressors. All comparisons involving eremus are at least moderately supported (Z > 2), but 30% and 70% of comparisons involving ingressus and spelaeus, respectively, fell below this threshold (indicated by open circles).
Fig. 3 |. Test of gene flow direction based on the distribution of rooted tree topologies along a non-overlapping 25 kb sliding window.
Five individuals are used in the test: a brown bear, a polar bear, the least admixed Caucasus cave bear (kudarensis, ‘cave –’), a more admixed European cave bear (spelaeus, ‘cave +’) and the Asiatic black bear outgroup. a, The symmetrical rooted ingroup species tree and the four alternative classes of topology that are informative on gene flow directionality. b, The proportion of 25 kb genomic blocks returning each topology class for eight brown bears, each represented by individual numbered bars: 1, Alaska (ABC Islands), North America; 2, Alaska (Denali), North America; 3, Russia; 4, Slovenia; 5, Sweden; 6, Georgia; 7, Spain; 8, Late Pleistocene Austria. The results support bidirectional gene flow among cave bears and brown bears: overrepresentation of topology class 1 relative to class 2 for all brown bears investigated indicates gene flow from cave bears into brown bears, and overrepresentation of topology class 3 relative to class 4 indicates gene flow in the opposite direction, from the brown/polar bear lineage into the more admixed cave bear. c, The cumulative size distributions of regions returning each topology class, determined by counting the number of consecutive 25 kb blocks returning the same topology. Absolute counts are provided in Supplementary Table 5. Individual points represent the cumulative proportions for individual brown bears. For the purpose of visualization, average cumulative proportions for each topology class are shown as coloured lines centred on the mean proportion observed for each block size and linked by linear interpolation.
Similar articles
- Ancient DNA reveals differences in behaviour and sociality between brown bears and extinct cave bears.
Fortes GG, Grandal-d'Anglade A, Kolbe B, Fernandes D, Meleg IN, García-Vázquez A, Pinto-Llona AC, Constantin S, de Torres TJ, Ortiz JE, Frischauf C, Rabeder G, Hofreiter M, Barlow A. Fortes GG, et al. Mol Ecol. 2016 Oct;25(19):4907-18. doi: 10.1111/mec.13800. Epub 2016 Sep 6. Mol Ecol. 2016. PMID: 27506329 - Genomic Evidence of Widespread Admixture from Polar Bears into Brown Bears during the Last Ice Age.
Cahill JA, Heintzman PD, Harris K, Teasdale MD, Kapp J, Soares AER, Stirling I, Bradley D, Edwards CJ, Graim K, Kisleika AA, Malev AV, Monaghan N, Green RE, Shapiro B. Cahill JA, et al. Mol Biol Evol. 2018 May 1;35(5):1120-1129. doi: 10.1093/molbev/msy018. Mol Biol Evol. 2018. PMID: 29471451 - Ancient DNA analyses reveal high mitochondrial DNA sequence diversity and parallel morphological evolution of late pleistocene cave bears.
Hofreiter M, Capelli C, Krings M, Waits L, Conard N, Münzel S, Rabeder G, Nagel D, Paunovic M, Jambrĕsić G, Meyer S, Weiss G, Pääbo S. Hofreiter M, et al. Mol Biol Evol. 2002 Aug;19(8):1244-50. doi: 10.1093/oxfordjournals.molbev.a004185. Mol Biol Evol. 2002. PMID: 12140236 - Impacts of Human Recreation on Brown Bears (Ursus arctos): A Review and New Management Tool.
Fortin JK, Rode KD, Hilderbrand GV, Wilder J, Farley S, Jorgensen C, Marcot BG. Fortin JK, et al. PLoS One. 2016 Jan 5;11(1):e0141983. doi: 10.1371/journal.pone.0141983. eCollection 2016. PLoS One. 2016. PMID: 26731652 Free PMC article. Review. - Sequencing and assembling bear genomes: the bare necessities.
Willey C, Korstanje R. Willey C, et al. Front Zool. 2022 Nov 30;19(1):30. doi: 10.1186/s12983-022-00475-8. Front Zool. 2022. PMID: 36451195 Free PMC article. Review.
Cited by
- The origins and diversification of Holarctic brown bear populations inferred from genomes of past and present populations.
Segawa T, Rey-Iglesia A, Lorenzen ED, Westbury MV. Segawa T, et al. Proc Biol Sci. 2024 Jan 31;291(2015):20232411. doi: 10.1098/rspb.2023.2411. Epub 2024 Jan 24. Proc Biol Sci. 2024. PMID: 38264778 - Population Genomic Analysis of North American Eastern Wolves (Canis lycaon) Supports Their Conservation Priority Status.
Heppenheimer E, Harrigan RJ, Rutledge LY, Koepfli KP, DeCandia AL, Brzeski KE, Benson JF, Wheeldon T, Patterson BR, Kays R, Hohenlohe PA, von Holdt BM. Heppenheimer E, et al. Genes (Basel). 2018 Dec 4;9(12):606. doi: 10.3390/genes9120606. Genes (Basel). 2018. PMID: 30518163 Free PMC article. - Demographic History of the Brown Bear (Ursus arctos) on Hokkaido Island, Japan, Based on Whole-Genomic Sequence Analysis.
Endo Y, Osada N, Mano T, Masuda R. Endo Y, et al. Genome Biol Evol. 2021 Sep 1;13(9):evab195. doi: 10.1093/gbe/evab195. Genome Biol Evol. 2021. PMID: 34410373 Free PMC article. - Historical biogeography of the leopard (Panthera pardus) and its extinct Eurasian populations.
Paijmans JLA, Barlow A, Förster DW, Henneberger K, Meyer M, Nickel B, Nagel D, Worsøe Havmøller R, Baryshnikov GF, Joger U, Rosendahl W, Hofreiter M. Paijmans JLA, et al. BMC Evol Biol. 2018 Oct 23;18(1):156. doi: 10.1186/s12862-018-1268-0. BMC Evol Biol. 2018. PMID: 30348080 Free PMC article. - Comparative analysis of microsatellites in coding regions provides insights into the adaptability of the giant panda, polar bear and brown bear.
Cheng M, Xie D, Price M, Zhou C, Zhang X. Cheng M, et al. Genetica. 2022 Dec;150(6):355-366. doi: 10.1007/s10709-022-00173-7. Epub 2022 Oct 26. Genetica. 2022. PMID: 36287311
References
- Shurtliff QR Mammalian hybrid zones: a review. Mamm. Rev. 43, 1–21 (2013).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous