A Briefly Argued Case That Asgard Archaea Are Part of the Eukaryote Tree - PubMed (original) (raw)
A Briefly Argued Case That Asgard Archaea Are Part of the Eukaryote Tree
Gregory P Fournier et al. Front Microbiol. 2018.
Abstract
The recent discovery of the Lokiarchaeota and other members of the Asgard superphylum suggests that closer analysis of the cell biology and evolution of these groups may help shed light on the origin of the eukaryote cell. Asgard lineages often appear in molecular phylogenies as closely related to eukaryotes, and possess "Eukaryote Signature Proteins" coded by genes previously thought to be unique to eukaryotes. This phylogenetic affinity to eukaryotes has been widely interpreted as indicating that Asgard lineages are "eukaryote-like archaea," with eukaryotes evolving from within a paraphyletic Archaea. Guided by the established principles of systematics, we examine the potential implications of the monophyly of Asgard lineages and Eukarya. We show that a helpful parallel case is that of Synapsida, a group that includes modern mammals and their more "reptile-like" ancestors, united by shared derived characters that evolved in their common ancestor. While this group contains extinct members that share many similarities with modern reptiles and their extinct relatives, they are evolutionarily distinct from Sauropsida, the group which includes modern birds, reptiles, and all other amniotes. Similarly, Asgard lineages and eukaryotes are united by shared derived characters to the exclusion of all other groups. Consequently, the Asgard group is not only highly informative for our understanding of eukaryogenesis, but may be better understood as being early diverging members of a broader group including eukaryotes, for which we propose the name "Eukaryomorpha." Significantly, this means that the relationship between Eukarya and Asgard lineages cannot, on its own, resolve the debate over 2 vs. 3 Domains of life; instead, resolving this debate depends upon identifying the root of Archaea with respect to Bacteria.
Keywords: Archaea; Asgard; Domains; Eukarya; cladistics; eukaryogenesis; synapomorphy; systematics.
Figures
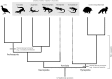
FIGURE 1
Phylogeny of selected major groups of amniotes. Selected synapomorphies are indicated by gray bars. Two basal extinct lineages Parareptilia and Archaeothyris (representing Ophiacodontidae) are included to indicate the retention of shared ancestral characters (large gray box). Sauropsida includes all extant reptiles and their extinct relatives. Extant anapsids (turtles) are omitted due to their uncertain placement within Sauropsida. Synapsida and Sauropsida are clades of equal taxonomic rank, with a last common ancestor congruent with the ancestor of all crown group amniotes (Amniota). Archosauria is a clade nested within Sauropsida. As such, it has a lower taxonomic rank than either Sauropsida or Synapsida, even if shared derived characters of this group distinguish them from other amniotes. Representative taxon images were downloaded from PhyloPic (
). All images are uncopyrighted except parareptilia (
http://phylopic.org/image/00b96cf3-1802-4bda-a6cc-76aea0f6f05e/
) which is owned by Nobu Tamura (vectorized by T. Michael Keesey), under the following creative commons license (
https://creativecommons.org/licenses/by-sa/3.0/legalcode
).
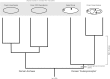
FIGURE 2
Hypothetical rooted archaeal tree including Asgard and eukaryotes, consistent with a “3-Domain” topology. ESPs (black dots) are synapomorphies uniting Asgard and eukaryotes as a clade (gray bar). Additional archaeal groups (e.g., DPANN) are omitted for clarity. Archaeal and Asgard groups share many “archaeal-like” ancestral characters (large gray box). Archaea are depicted as rooted between Euryarchaeota and TACK, although this hypothetical 3-Domain topology is also valid under all other rootings for this group. Despite these ancestral characters, with this rooting, Asgard + Eukarya together constitute a clade of equal taxonomic rank to that of Archaea. Here, we propose the name “Eukaryomorpha,” reflecting the shared characters uniting this group and distinguishing them from the archaeal outgroup.
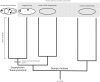
FIGURE 3
Hypothetical rooted archaeal tree including Asgard and eukaryotes, consistent with a “2-Domain” topology. With this rooting, the Asgard + Eukarya clade is nested within Archaea, i.e., the last common ancestor of all Archaea (excluding Asgard) is also the last common ancestor of Eukarya and Asgard. As such, Asgard + Eukarya constitute a clade of unequal taxonomic rank to Archaea. Note that the scenarios in Figures 2, 3 only differ in the placement of the root, not in the topology of the tree or mapping of the characters.
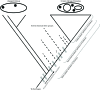
FIGURE 4
Cladogram of Asgard and eukaryote evolution as related to different definitions of Eukarya. Eukaryal-specific characters (ESPs, gray boxes) were acquired in the ancestor of Asgard + Eukarya, and continue to be acquired in the eukaryal stem lineage. An apomorphy-based definition of Eukarya requires the identification of a specific defining character for Eukarya, which would include some eukaryal stem groups, but exclude others. The selection of a defining character for an apomorphy-based definition is therefore inherently subjective. The Asgard group is depicted as an evolutionary grade, although a monophyletic Asgard group is also consistent with this model.
Similar articles
- Coevolution of Eukaryote-like Vps4 and ESCRT-III Subunits in the Asgard Archaea.
Lu Z, Fu T, Li T, Liu Y, Zhang S, Li J, Dai J, Koonin EV, Li G, Chu H, Li M. Lu Z, et al. mBio. 2020 May 19;11(3):e00417-20. doi: 10.1128/mBio.00417-20. mBio. 2020. PMID: 32430468 Free PMC article. - The Asgard Archaeal-Unique Contribution to Protein Families of the Eukaryotic Common Ancestor Was 0.3.
Knopp M, Stockhorst S, van der Giezen M, Garg SG, Gould SB. Knopp M, et al. Genome Biol Evol. 2021 Jun 8;13(6):evab085. doi: 10.1093/gbe/evab085. Genome Biol Evol. 2021. PMID: 33892498 Free PMC article. - The expanding Asgard archaea and their elusive relationships with Eukarya.
Da Cunha V, Gaïa M, Forterre P. Da Cunha V, et al. mLife. 2022 Mar 24;1(1):3-12. doi: 10.1002/mlf2.12012. eCollection 2022 Mar. mLife. 2022. PMID: 38818326 Free PMC article. - Asgard archaea: Diversity, function, and evolutionary implications in a range of microbiomes.
MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP. MacLeod F, et al. AIMS Microbiol. 2019 Jan 30;5(1):48-61. doi: 10.3934/microbiol.2019.1.48. eCollection 2019. AIMS Microbiol. 2019. PMID: 31384702 Free PMC article. Review. - [Progress in elucidating the origin of eukaryotes].
Gao ZW, Wang L. Gao ZW, et al. Yi Chuan. 2020 Oct 20;42(10):929-948. doi: 10.16288/j.yczz.20-107. Yi Chuan. 2020. PMID: 33229320 Review. Chinese.
Cited by
- Eukaryotes Are a Holophyletic Group of Polyphyletic Origin.
Skejo J, Franjević D. Skejo J, et al. Front Microbiol. 2020 Jul 2;11:1380. doi: 10.3389/fmicb.2020.01380. eCollection 2020. Front Microbiol. 2020. PMID: 32714303 Free PMC article. No abstract available. - Insights into the evolution of enzymatic specificity and catalysis: From Asgard archaea to human adenylate kinases.
Verma A, Åberg-Zingmark E, Sparrman T, Mushtaq AU, Rogne P, Grundström C, Berntsson R, Sauer UH, Backman L, Nam K, Sauer-Eriksson E, Wolf-Watz M. Verma A, et al. Sci Adv. 2022 Nov 4;8(44):eabm4089. doi: 10.1126/sciadv.abm4089. Epub 2022 Nov 4. Sci Adv. 2022. PMID: 36332013 Free PMC article. - Manifold Routes to a Nucleus.
Hendrickson HL, Poole AM. Hendrickson HL, et al. Front Microbiol. 2018 Oct 26;9:2604. doi: 10.3389/fmicb.2018.02604. eCollection 2018. Front Microbiol. 2018. PMID: 30416499 Free PMC article. - The catalytic and structural basis of archaeal glycerophospholipid biosynthesis.
de Kok NAW, Driessen AJM. de Kok NAW, et al. Extremophiles. 2022 Aug 17;26(3):29. doi: 10.1007/s00792-022-01277-w. Extremophiles. 2022. PMID: 35976526 Free PMC article. Review. - Division of labour in a matrix, rather than phagocytosis or endosymbiosis, as a route for the origin of eukaryotic cells.
Bateman A. Bateman A. Biol Direct. 2020 Apr 28;15(1):8. doi: 10.1186/s13062-020-00260-9. Biol Direct. 2020. PMID: 32345370 Free PMC article.
References
- Benson R. B. J. (2012). Interrelationships of basal synapsids: cranial and postcranial morphological partitions suggest different topologies. J. Syst. Palaeontol. 10 601–624. 10.1080/14772019.2011.631042 - DOI
- Benton M. J. (2008). Classification and phylogeny of the diapsid reptiles. Zool. J. Linn. Soc. 84 97–164. 10.1111/j.1096-3642.1985.tb01796.x - DOI
- Benton M. J. (2015). Vertebrate Palaeontology. Hoboken, NJ: Wiley Blackwell.
LinkOut - more resources
Full Text Sources
Other Literature Sources