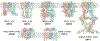Single-particle cryo-EM-How did it get here and where will it go - PubMed (original) (raw)
Review
Single-particle cryo-EM-How did it get here and where will it go
Yifan Cheng. Science. 2018.
Abstract
Cryo-electron microscopy, or simply cryo-EM, refers mainly to three very different yet closely related techniques: electron crystallography, single-particle cryo-EM, and electron cryotomography. In the past few years, single-particle cryo-EM in particular has triggered a revolution in structural biology and has become a newly dominant discipline. This Review examines the fascinating story of its start and evolution over the past 40-plus years, delves into how and why the recent technological advances have been so groundbreaking, and briefly considers where the technique may be headed in the future.
Copyright © 2018, American Association for the Advancement of Science.
Figures
Figure 1.. Establishing of single particle cryo-EM
A. An electron diffraction pattern of frozen hydrated catalase crystal (9). The diffraction spots are visible at beyond 3 Å resolution. This experiment established the concept of cryo-EM. B. An electron micrograph of frozen hydrated adenovirus particle recorded from a frozen hydrated grid prepared by plunge freezing. The micrograph is from the same dataset described in (11) and reproduced from (9). C. The first 3D reconstruction of bacteriorhodopsin determined by electron crystallography (15). D. A 3D model of the 50S ribosome subunit determined by single particle reconstruction of negatively stained large ribosomal subunit from Escherichia coli (22).
Figure 2.. Direct electron detection camera enabled atomic structure determination
A. Comparison of DQE curves of a scintillator based CCD camera (black), and direct electron detection camera K2 operating in base mode (blue) and super-resolution counting mode (red) (31). B. A typical electron micrograph of archaeal 20S proteasome (~700kDa in molecular weight) recorded using direct electron detection camera (60). C. Fourier power spectrum calculated from the image (B) (60). High-resolution signal at ~3Å resolution is clearly visible (60). D. A portion of density map from the 3D reconstruction of archaeel 20S proteasome (31).
Figure 3.. Single particle cryo-EM enables atomic structure determination of TRP channels
Ribbon diagrams of atomic structures from each subfamily of TRP channel superfamily. They are TRPV1 (49), TRPA1 (61), TRPM4 (62), TRPC4 (63), NOMOPC (also named TRPN1) (64), PKD2 (or TRPP2) (65) and TRPML (66). The rapid pace of integral membrane protein structure determination is enabled by single particle cryo-EM and is unprecedented.
Similar articles
- Structure Determination by Single-Particle Cryo-Electron Microscopy: Only the Sky (and Intrinsic Disorder) is the Limit.
Nwanochie E, Uversky VN. Nwanochie E, et al. Int J Mol Sci. 2019 Aug 27;20(17):4186. doi: 10.3390/ijms20174186. Int J Mol Sci. 2019. PMID: 31461845 Free PMC article. Review. - How cryo-electron microscopy and X-ray crystallography complement each other.
Wang HW, Wang JW. Wang HW, et al. Protein Sci. 2017 Jan;26(1):32-39. doi: 10.1002/pro.3022. Epub 2016 Sep 7. Protein Sci. 2017. PMID: 27543495 Free PMC article. Review. - Dawning of a new era in TRP channel structural biology by cryo-electron microscopy.
Madej MG, Ziegler CM. Madej MG, et al. Pflugers Arch. 2018 Feb;470(2):213-225. doi: 10.1007/s00424-018-2107-2. Epub 2018 Jan 17. Pflugers Arch. 2018. PMID: 29344776 Review. - A Super-Clustering Approach for Fully Automated Single Particle Picking in Cryo-EM.
Al-Azzawi A, Ouadou A, Tanner JJ, Cheng J. Al-Azzawi A, et al. Genes (Basel). 2019 Aug 30;10(9):666. doi: 10.3390/genes10090666. Genes (Basel). 2019. PMID: 31480377 Free PMC article. - Single-Particle Cryo-EM of Membrane Proteins in Lipid Nanodiscs.
Kalienkova V, Alvadia C, Clerico Mosina V, Paulino C. Kalienkova V, et al. Methods Mol Biol. 2020;2127:245-273. doi: 10.1007/978-1-0716-0373-4_17. Methods Mol Biol. 2020. PMID: 32112327
Cited by
- Structures of the Mononegavirales Polymerases.
Liang B. Liang B. J Virol. 2020 Oct 27;94(22):e00175-20. doi: 10.1128/JVI.00175-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847861 Free PMC article. Review. - Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS).
Drew D, North RA, Nagarathinam K, Tanabe M. Drew D, et al. Chem Rev. 2021 May 12;121(9):5289-5335. doi: 10.1021/acs.chemrev.0c00983. Epub 2021 Apr 22. Chem Rev. 2021. PMID: 33886296 Free PMC article. - Changing Form and Function through Carotenoids and Synthetic Biology.
Wurtzel ET. Wurtzel ET. Plant Physiol. 2019 Mar;179(3):830-843. doi: 10.1104/pp.18.01122. Epub 2018 Oct 25. Plant Physiol. 2019. PMID: 30361256 Free PMC article. - Effects of cryo-EM cooling on structural ensembles.
Bock LV, Grubmüller H. Bock LV, et al. Nat Commun. 2022 Mar 31;13(1):1709. doi: 10.1038/s41467-022-29332-2. Nat Commun. 2022. PMID: 35361752 Free PMC article. - Sequence and structural conservation reveal fingerprint residues in TRP channels.
Cabezas-Bratesco D, Mcgee FA, Colenso CK, Zavala K, Granata D, Carnevale V, Opazo JC, Brauchi SE. Cabezas-Bratesco D, et al. Elife. 2022 Jun 10;11:e73645. doi: 10.7554/eLife.73645. Elife. 2022. PMID: 35686986 Free PMC article.
References
- Feynman RP, in Feynman and computation, Hey AJG, Ed. (Perseus Books Cambridge, MA, USA, 1999), pp. 63–76.
- Shi Y, A glimpse of structural biology through X-ray crystallography. Cell 159, 995–1014 (2014). - PubMed
- Jones N, Crystallography: Atomic secrets. Nature 505, 602–603 (2014). - PubMed
- Frank J, Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy 1, 159–162 (1975). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM082893/GM/NIGMS NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- S10 OD021741/OD/NIH HHS/United States
- R01 GM098672/GM/NIGMS NIH HHS/United States
- S10 OD020054/OD/NIH HHS/United States
- R01 HL134183/HL/NHLBI NIH HHS/United States
- P01 GM111126/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous