Chemical proteomics reveals new targets of cysteine sulfinic acid reductase - PubMed (original) (raw)
Chemical proteomics reveals new targets of cysteine sulfinic acid reductase
Salma Akter et al. Nat Chem Biol. 2018 Nov.
Abstract
Cysteine sulfinic acid or S-sulfinylation is an oxidative post-translational modification (OxiPTM) that is known to be involved in redox-dependent regulation of protein function but has been historically difficult to analyze biochemically. To facilitate the detection of S-sulfinylated proteins, we demonstrate that a clickable, electrophilic diazene probe (DiaAlk) enables capture and site-centric proteomic analysis of this OxiPTM. Using this workflow, we revealed a striking difference between sulfenic acid modification (S-sulfenylation) and the S-sulfinylation dynamic response to oxidative stress, which is indicative of different roles for these OxiPTMs in redox regulation. We also identified >55 heretofore-unknown protein substrates of the cysteine sulfinic acid reductase sulfiredoxin, extending its function well beyond those of 2-cysteine peroxiredoxins (2-Cys PRDX1-4) and offering new insights into the role of this unique oxidoreductase as a central mediator of reactive oxygen species-associated diseases, particularly cancer. DiaAlk therefore provides a novel tool to profile S-sulfinylated proteins and study their regulatory mechanisms in cells.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
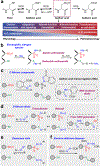
Figure 1 |. Development of electrophilic nitrogen species (ENS) for labeling sulfinic acids.
(a) The sulfur atom of cysteine has the ability to assume many different oxidation states. As the balance between H2O2 production and catabolism changes, adaption occurs with subsequent activation of signaling pathways via protein cysteine OxiPTM, which may be reversible or irreversible. An excessive imbalance can lead to cellular transformation or cell death. (b) General ENS approach for labeling of sulfinic acid in the presence of thiols. (c) Sulfinic acid nitroso ligation (SNL). (d) Reactivity of sulfinic acids and thiols with _S_-nitrosothiols. (e) Diazonium salts react with both sulfinic acids and thiols, generating a reversible adduct. (f) Electron-deficient diazenes promote covalent modification of sulfinic acids to give a stable sulfonamide adduct.
Figure 2 |. Reactivity of electron-deficient diazenes toward recombinant protein models.
(a) Labeling efficiency of electron-deficient diazenes with recombinant C64,82S Gpx3-SO2H as measured by intact protein MS. (b) Synthesis of DiaAlk (5). (c) Sulfinic acid attacks the less hindered nitrogen of the diazene to generate the initial sulfonamide adduct (+ 298 Da). The BOC group undergoes hydrolysis to yield the final product (+ 198 Da). (d) Deconvoluted MS spectra of C64,82S Gpx3-SO2H before and after treatment with Dia-Alk at various times and pH. (e) ESI-MS spectra of C64,82S Gpx3-SH before and after Dia-Alk treatment, followed by DTT. (f-h) Immunoblot showing DiaAlk detection of sulfinic acid in C64,82S Gpx3 (f), DJ-1 (g) and PspE (h) by streptavidin–HRP. Equal protein loading was confirmed by re-probing immunoblots with the indicated antibody. Representative data from three independent experiments are shown. Uncropped scans of immunoblots are provided in Supplementary Fig. 22.
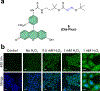
Figure 3 |. Detection of protein _S_-sulfinylation from cells using DiaFluo.
(a) Structure of fluorescein-tagged DiaAlk analog, DiaFluo (6). (b) Fluorescence microscopy images of _S_-sulfinylation in HeLa cells before and after stimulation with H2O2. Cells were fixed, permeabilized, and blocked with NEM. Protein _S_-sulfinylation was visualized at 488 nm (green) after treatment with or without (control) DiaFluo. Nuclei were counterstained with DAPI (blue). Scale bars, 20 μm. Representative data from three independent experiments and more than 40 images are shown.
Figure 4 |. Site-centric and quantitative chemoproteomic profiling of protein _S_-sulfinylation.
(a) Schematic representation of site-centric quantitative chemoproteomic workflow for global profiling of _S_-sulfinylation in native proteomes. (b) Characteristic fragmentation of DiaAlk-triazohexanoic acid modified peptides. DFI: diagnostic fragment ions. (c) MS/MS spectrum of a DiaAlk-tagged peptide, which unequivocally identifies Cys47 of PRDX6 as a _S_-sulfinylated site. (d) Site-specific changes in _S_-sulfinylation in A549 (top) and HeLa (bottom) cells in response to H2O2 stress. Heavy (H2O2) to light (control) ratios are correlated with functional annotations from the UniProt database. (e) Venn diagram of A549- and HeLa-sulfinylomes labeled with DiaAlk. (f) Extracted ion chromatograms showing changes in DiaAlk-tagged peptides from PRDX6 (Cys47), GAPDH (Cys152), GSTO1 (Cys192), and PTPN1 (Cys215) from H2O2 stimulation of A549 (left) and HeLa cells (right). The profiles for light- and heavy-labeled peptides are shown in red and blue, respectively. Heavy (H2O2) to light (control) ratios were calculated from three independent experiments and are displayed below the individual chromatograms. (g) DiaAlk labels HA-tagged wild-type PTPN1, but not C215S, in HeLa cells. Representative data from three independent experiments are shown. Uncropped scans of immunoblots are provided in Supplementary Fig. 22.
Figure 5 |. Comparison of _S_-sulfenylome and _S_-sulfinylome sites and dynamic fold-changes.
(a) Venn diagram showing overlap between the _S_-sulfenylome and _S_-sulfinylome. Ratios (H2O2 versus untreated control) obtained from common sites in the _S_-sulfenylome and _S_-sulfinylome dataset are plotted as line series and displayed on a log10 scale on the y-axis. Two-fold or smaller changes between –SOH and –SO2H are depicted with light grey lines; fold-changes greater than two are depicted in black. (b) Ratio (H2O2 versus untreated control) of _S_-sulfenylated and _S_-sulfinylated sites identified in the same cell line are plotted as a scatter graph and displayed on a log10 scale on the y-axis.
Figure 6 |. Proteome-wide analysis of SRX-regulated changes in _S_-sulfinylation.
(a) Chemoproteomic workflow to map differential _S_-sulfinylation in SRX deficient (Srx−/−) and SRX replete (Srx+/+) MEFs, classified as “SRX-regulated” cysteines. (b) Venn diagram showing overlap between the Srx+/+ and _Srx−/− S_-sulfinylomes in MEFs. (c) Distribution of dynamic (measured ratios) of protein _S_-sulfinylation in Srx+/+ (blue) and Srx−/− (red) MEFs. Ratios between heavy (recovery) and light (without recovery, or control) were measured as the turnover rates for each _S_-sulfinylation event. Inset, extracted ion chromatograms (XIC) of DiaAlk-modified peptides from PRDX6 (Cys47) and GAPDH (Cys152) from Srx +/+ MEFs. The profiles for light- and heavy-labeled peptides are shown in red and blue, respectively. Heavy (recovery) to light (control) ratios were calculated from two independent experiments and are displayed below the individual chromatograms. (d) _S_-sulfinylation of PRDX1, but not GAPDH, is reversible and _Srx_-dependent, as detected by DiaAlk. Representative data from three independent experiments are shown. Uncropped scans of immunoblots are provided in Supplementary Fig. 22. (e) Line series plot showing the turnover rates of the same _S_-sulfinylation sites identified in both Srx+/+ and Srx−/− MEFs. (f,g) XIC of DiaAlk-modified peptides from PTPN12 (Cys164) (f) and DJ-1 (Cys46) (g) from Srx+/+ and Srx−/− MEFs. Heavy (recovery) to light (control) ratios were calculated from two independent experiments and are displayed below the individual chromatograms. (h-j) Recombinant SRX1 reduces _S_-sulfinylation of PTPN12 (h) or DJ-1 (i), but not GAPDH (j), as detected by BioDiaAlk (7). Uncropped scans of immunoblots are provided in Supplementary Fig. 22. (k) Luminescence assay measures SRX1 ATPase activity in the presence of PTPN12 or DJ-1. Signal in the absence of SRX1 was subtracted as background; control reaction lacks ATP. Representative data from three independent experiments are shown, each performed in triplicate; error bars represent the standard deviation (s.d).
Comment in
- Finding S-sulfinylated proteins.
Doerr A. Doerr A. Nat Methods. 2018 Nov;15(11):859. doi: 10.1038/s41592-018-0200-2. Nat Methods. 2018. PMID: 30377350 No abstract available.
Similar articles
- Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation.
Shi Y, Carroll KS. Shi Y, et al. Acc Chem Res. 2020 Jan 21;53(1):20-31. doi: 10.1021/acs.accounts.9b00562. Epub 2019 Dec 23. Acc Chem Res. 2020. PMID: 31869209 Free PMC article. Review. - Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine.
Chang TS, Jeong W, Woo HA, Lee SM, Park S, Rhee SG. Chang TS, et al. J Biol Chem. 2004 Dec 3;279(49):50994-1001. doi: 10.1074/jbc.M409482200. Epub 2004 Sep 24. J Biol Chem. 2004. PMID: 15448164 - Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins.
Woo HA, Jeong W, Chang TS, Park KJ, Park SJ, Yang JS, Rhee SG. Woo HA, et al. J Biol Chem. 2005 Feb 4;280(5):3125-8. doi: 10.1074/jbc.C400496200. Epub 2004 Dec 8. J Biol Chem. 2005. PMID: 15590625 - The redox biochemistry of protein sulfenylation and sulfinylation.
Lo Conte M, Carroll KS. Lo Conte M, et al. J Biol Chem. 2013 Sep 13;288(37):26480-8. doi: 10.1074/jbc.R113.467738. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861405 Free PMC article. Review. - A Chemical Approach for the Detection of Protein Sulfinylation.
Lo Conte M, Lin J, Wilson MA, Carroll KS. Lo Conte M, et al. ACS Chem Biol. 2015 Aug 21;10(8):1825-30. doi: 10.1021/acschembio.5b00124. Epub 2015 Jun 17. ACS Chem Biol. 2015. PMID: 26039147 Free PMC article.
Cited by
- Protein Oxidative Modifications in Neurodegenerative Diseases: From Advances in Detection and Modelling to Their Use as Disease Biomarkers.
Anjo SI, He Z, Hussain Z, Farooq A, McIntyre A, Laughton CA, Carvalho AN, Finelli MJ. Anjo SI, et al. Antioxidants (Basel). 2024 May 31;13(6):681. doi: 10.3390/antiox13060681. Antioxidants (Basel). 2024. PMID: 38929122 Free PMC article. Review. - Hyperoxidation of Peroxiredoxins and Effects on Physiology of Drosophila.
McGinnis A, Klichko VI, Orr WC, Radyuk SN. McGinnis A, et al. Antioxidants (Basel). 2021 Apr 15;10(4):606. doi: 10.3390/antiox10040606. Antioxidants (Basel). 2021. PMID: 33920774 Free PMC article. - Metabolic Hallmarks of Hepatic Stellate Cells in Liver Fibrosis.
Khomich O, Ivanov AV, Bartosch B. Khomich O, et al. Cells. 2019 Dec 20;9(1):24. doi: 10.3390/cells9010024. Cells. 2019. PMID: 31861818 Free PMC article. Review. - Redox Systems Biology: Harnessing the Sentinels of the Cysteine Redoxome.
Held JM. Held JM. Antioxid Redox Signal. 2020 Apr 1;32(10):659-676. doi: 10.1089/ars.2019.7725. Epub 2019 Sep 9. Antioxid Redox Signal. 2020. PMID: 31368359 Free PMC article. Review. - Detection, identification, and quantification of oxidative protein modifications.
Hawkins CL, Davies MJ. Hawkins CL, et al. J Biol Chem. 2019 Dec 20;294(51):19683-19708. doi: 10.1074/jbc.REV119.006217. Epub 2019 Oct 31. J Biol Chem. 2019. PMID: 31672919 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA227849/CA/NCI NIH HHS/United States
- R01 CA174864/CA/NCI NIH HHS/United States
- R01 GM102187/GM/NIGMS NIH HHS/United States
- R01 GM072866/GM/NIGMS NIH HHS/United States
- P30 CA012197/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous