Revealing the impact of the Caucasus region on the genetic legacy of Romani people from genome-wide data - PubMed (original) (raw)
Revealing the impact of the Caucasus region on the genetic legacy of Romani people from genome-wide data
Zsolt Bánfai et al. PLoS One. 2018.
Abstract
Romani people are a significant minority in Europe counting about 10 million individuals scattered throughout the continent. They are a migratory group originating from Northwestern India. Their exodus from India occurred approximately 1000-1500 years ago. The migration route of the Romani people was reconstructed with the help of cultural anthropology, linguistics and historical records. Their migration made them through Central Asia, Middle East and the Caucasus region, prior to the arriving into Europe. Yet the significance of these regions, especially of the Caucasus, in Roma ancestry was a rather neglected topic. Contribution of the Caucasus and further affected regions to the ancestry of Roma was investigated based on genome-wide autosomal marker data. 158 European Roma samples and 41 populations from the Caucasus region, from Middle East, Central Asia and from South Asia were considered in our tests. Population structure and ancestry analysis algorithms were applied to investigate the relationship of Roma with these populations. Identical by descent DNA segment analyses and admixture linkage disequilibrium based tests were also applied. Our results suggest that the Caucasus region plays also a significant role in the genetic legacy of Romani people besides the main sources, Europe and South Asia, previously investigated by other population genetic studies. The Middle East and Central Asia seems slightly less important but far from negligible in connection with the sources of Roma ancestry. Our results point out that the Caucasus region and altogether the area of the Caspian and Black Seas had a significant role in the migration of Romani people towards Europe and contributed significantly to the genetic legacy of Roma rival to the European and Indian main sources.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
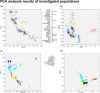
Fig 1. PCA analysis results featuring all populations.
Each symbol represents an individual. (a) Shows the population structure of investigated major regional populations on principal components 1 and 2. (b) Shows the population structure of major populations on principal component 2 and 4. (c) All populations included in PCA grouped into regions on principal component 1 and 2 (d) Structure of all populations included in the PCA plotted on principal component 3 and 4. Note that all four graphs are the result of the same PCA. Eigenvalues of PC1 and PC2 were 19.38 and 6.67, eigenvalues of PC3 and PC4 were 6.07 and 5.31.
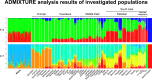
Fig 2. ADMIXTURE analysis results at K = 3 and 7 hypothetical ancestral groups.
Each column group represents one population, each column represents one individual. The number of individuals in this analysis was restricted to a maximum size of 30, except of Roma. A figure containing all ADMIXTURE graphs from K = 3 to K = 10 can be found in the supplemental material (S1 Fig).
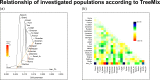
Fig 3. TreeMix analysis results.
(a) Maximum likelihood graph and 3 included migration events estimated by TreeMix. Contains population from all investigated regions. (b) The residual fit from the ML graph.
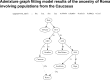
Fig 4. Model setup of the proposed admixture between Roma ancestors and Caucasus populations tested with Admixture graph fitting.
Branch lengths are shown in units of Fst*1000. Worst F-statistics result is shown above the graph. OoA—Out of Africa, AWE—Ancestral West Eurasians, AEE—Ancestral East Eurasians, ASA—Ancestral South Asians.
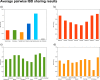
Fig 5. Average pairwise IBD sharing between Roma and the investigated regional populations.
(a) IBD share between Roma and investigated regions. (b) IBD share between Roma and South Asian populations. (c) IBD share of Roma with the Caucasus. (d) IBD share of Roma with Middle East and Central Asian populations.
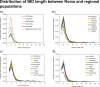
Fig 6. Average IBD length distribution between Roma and all investigated regions.
(a) Average IBD length distribution between Roma and all regions. (b) Average IBD length distribution of Roma with South Asian groups. (c) Average IBD length distribution of Roma with the Caucasus. (d) Average IBD length distribution of Roma with populations from the Middle East and Central Asia.
Similar articles
- Population Genetics of the European Roma-A Review.
Ena GF, Aizpurua-Iraola J, Font-Porterias N, Calafell F, Comas D. Ena GF, et al. Genes (Basel). 2022 Nov 8;13(11):2068. doi: 10.3390/genes13112068. Genes (Basel). 2022. PMID: 36360305 Free PMC article. Review. - Reconstructing Roma history from genome-wide data.
Moorjani P, Patterson N, Loh PR, Lipson M, Kisfali P, Melegh BI, Bonin M, Kádaši L, Rieß O, Berger B, Reich D, Melegh B. Moorjani P, et al. PLoS One. 2013;8(3):e58633. doi: 10.1371/journal.pone.0058633. Epub 2013 Mar 13. PLoS One. 2013. PMID: 23516520 Free PMC article. - The role of the Vlax Roma in shaping the European Romani maternal genetic history.
Salihović MP, Barešić A, Klarić IM, Cukrov S, Lauc LB, Janićijević B. Salihović MP, et al. Am J Phys Anthropol. 2011 Oct;146(2):262-70. doi: 10.1002/ajpa.21566. Epub 2011 Aug 16. Am J Phys Anthropol. 2011. PMID: 21915846 - Refining the South Asian Origin of the Romani people.
Melegh BI, Banfai Z, Hadzsiev K, Miseta A, Melegh B. Melegh BI, et al. BMC Genet. 2017 Aug 31;18(1):82. doi: 10.1186/s12863-017-0547-x. BMC Genet. 2017. PMID: 28859608 Free PMC article. - Human genomic diversity in Europe: a summary of recent research and prospects for the future.
Cavalli-Sforza LL, Piazza A. Cavalli-Sforza LL, et al. Eur J Hum Genet. 1993;1(1):3-18. doi: 10.1159/000472383. Eur J Hum Genet. 1993. PMID: 7520820 Review.
Cited by
- European Roma groups show complex West Eurasian admixture footprints and a common South Asian genetic origin.
Font-Porterias N, Arauna LR, Poveda A, Bianco E, Rebato E, Prata MJ, Calafell F, Comas D. Font-Porterias N, et al. PLoS Genet. 2019 Sep 23;15(9):e1008417. doi: 10.1371/journal.pgen.1008417. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31545809 Free PMC article. - Population Genetics of the European Roma-A Review.
Ena GF, Aizpurua-Iraola J, Font-Porterias N, Calafell F, Comas D. Ena GF, et al. Genes (Basel). 2022 Nov 8;13(11):2068. doi: 10.3390/genes13112068. Genes (Basel). 2022. PMID: 36360305 Free PMC article. Review. - Genomic Insights into the Population History of the Resande or Swedish Travelers.
Vilà-Valls L, Aizpurua-Iraola J, Casinge S, Bojs K, Flores-Bello A, Font-Porterias N, Comas D. Vilà-Valls L, et al. Genome Biol Evol. 2023 Feb 3;15(2):evad006. doi: 10.1093/gbe/evad006. Genome Biol Evol. 2023. PMID: 36655389 Free PMC article. - Founder lineages in the Iberian Roma mitogenomes recapitulate the Roma diaspora and show the effects of demographic bottlenecks.
Aizpurua-Iraola J, Giménez A, Carballo-Mesa A, Calafell F, Comas D. Aizpurua-Iraola J, et al. Sci Rep. 2022 Nov 4;12(1):18720. doi: 10.1038/s41598-022-23349-9. Sci Rep. 2022. PMID: 36333436 Free PMC article.
References
- Liégeois JP. Roma, gypsies, travellers Strasbourg, France: Council of Europe; 1994.
- Marushiakova E, Popov V. Gypsies (Roma) in Bulgaria. Frankfurt, Germany: P. Lang; 1997.
- Magocsi P. Historical Atlas of East Central Europe; Cartographic design by Geoffrey J. Matthews: Seattle: University of Washington Press; 1993.
- Iovita RP, Schurr TG. Reconstructing the origins and migrations of diasporic populations: The case of the European gypsies. Am Anthropol. 2004;106(2):267–81. 10.1525/aa.2004.106.2.267 WOS:000222581500004. - DOI
- Fraser AM. The gypsies Oxford, UK: Blackwell Publishing; 1995.
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by GINOP-2.3.3-15-2016-00025; https://www.palyazat.gov.hu/evaluation; Research University Resource, Institutional Excellence Grant 2016, Centre for Excellence EFOP-3.6.3-VEKOP-16-2017-00009 - Centre of Molecular Medicine, Grant Manager: Ministry of Human Resources, Hungary and by the National Scientific Research Program (NKFI) K 119540. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources