Contradictory mRNA and protein misexpression of EEF1A1 in ductal breast carcinoma due to cell cycle regulation and cellular stress - PubMed (original) (raw)
Contradictory mRNA and protein misexpression of EEF1A1 in ductal breast carcinoma due to cell cycle regulation and cellular stress
Cheng-Yu Lin et al. Sci Rep. 2018.
Abstract
Encoded by EEF1A1, the eukaryotic translation elongation factor eEF1α1 strongly promotes the heat shock response, which protects cancer cells from proteotoxic stress, following for instance oxidative stress, hypoxia or aneuploidy. Unexpectedly, therefore, we find that EEF1A1 mRNA levels are reduced in virtually all breast cancers, in particular in ductal carcinomas. Univariate and multivariate analyses indicate that EEF1A1 mRNA underexpression independently predicts poor patient prognosis for estrogen receptor-positive (ER+) cancers. EEF1A1 mRNA levels are lowest in the most invasive, lymph node-positive, advanced stage and postmenopausal tumors. In sharp contrast, immunohistochemistry on 100 ductal breast carcinomas revealed that at the protein level eEF1α1 is ubiquitously overexpressed, especially in ER+ , progesterone receptor-positive and lymph node-negative tumors. Explaining this paradox, we find that EEF1A1 mRNA levels in breast carcinomas are low due to EEF1A1 allelic copy number loss, found in 27% of tumors, and cell cycle-specific expression, because mRNA levels are high in G1 and low in proliferating cells. This also links estrogen-induced cell proliferation to clinical observations. In contrast, high eEF1α1 protein levels protect tumor cells from stress-induced cell death. These observations suggest that, by obviating EEF1A1 transcription, cancer cells can rapidly induce the heat shock response following proteotoxic stress, and survive.
Conflict of interest statement
The authors declare no competing interests.
Figures
Figure 1
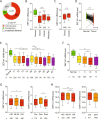
EEF1A1 mRNA is underexpressed in breast cancer. (A) EEF1A1 mRNA levels in normal breast and breast cancer were pairwise compared in 37 microarray studies. The graph shows the distribution of studies reporting underexpression or overexpression in tumors or significant differential expression, as indicated. P-value: Chi-square test. (B) Box plot of normalized EEF1A1 mRNA expression in indicated breast carcinomas compared to normal breast tissue using the TCGA breast cancer RNAseq dataset. Whiskers show 10–90 percentiles. P-values: Mann-Whitney U tests. (C) Box plot of normalized EEF1A1 mRNA expression in breast ductal carcinoma compared to lobular carcinoma using the METABRIC dataset. Whiskers show 10–90 percentiles. P-values: Mann-Whitney U test. (D) Normalized EEF1A1 mRNA expression in matched breast carcinoma and normal breast tissue pairs using the TCGA RNAseq dataset. P-value: Wilcoxon matched-pairs signed rank test. (E) Box plot of normalized EEF1A1 mRNA expression in breast tumors for TNM stages, which includes tumor invasion (T1-T4), nodal status (N0-N3) and metastatic states (M0-M1), as indicated. Whiskers show 10–90 percentiles. P-values: Mann-Whitney U tests. (F) Box plot showing normalized EEF1A1 mRNA expression in breast carcinomas for indicated tumor stages. Whiskers show 10–90 percentiles. P-values: Mann-Whitney U tests. (G) Box plot showing normalized EEF1A1 mRNA expression in breast tumors for age and menopausal status using the TCGA RNAseq dataset. Whiskers show 10–90 percentiles. P-values: Mann-Whitney U tests. (H) Box plot showing normalized EEF1A1 mRNA expression in breast tumors for age and menopausal status using the METABRIC dataset. Whiskers show 10–90 percentiles. P-values: Mann-Whitney U test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Figure 2
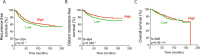
Low EEF1A1 mRNA expression in breast cancer predicts poor patient survival. (A) Recurrence-free survival curve of patients from the Kaplan-Meier plotter dataset. Patients were split into high and low EEF1A1 mRNA expression groups using median expression level as the cut-off, determined as previously described. (B) Distant metastasis-free survival curve. (C) Overall survival curve. P-values: log-rank Mantel-Cox tests. N/s, not significant; *p < 0.05.
Figure 3
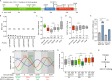
Low EEF1A1 mRNA expression in breast carcinoma is due to EEF1A1 allelic copy number loss and cell cycle-associated expression. (A) Mutations identified in 2,446 breast cancer samples from the COSMIC database (version 83, see Methods). The image was generated as described, and modified. Scale bar indicates amino acid numbers. (B) Box plot of β-values of the six CpG probes in the EEF1A1 promoter 1000 base pairs upstream of the EEF1A1 transcription start site in normal breast tissue (N) and breast tumor (T). Data are derived from TCGA. P-values: Mann-Whitney U test; n/s, not significant. (C) Box plot of EEF1A1 mRNA expression level in breast carcinomas and normal breast tissue for indicated EEF1A1 allelic copy number status. Data are derived from the TCGA RNAseq and SNP6 microarray datasets. P-values: Mann-Whitney U test. (D) Box plot as in (B) but using data from the METABRIC datasets,. P-values: Mann-Whitney U test. N/s, not significant; ****p < 0.0001. (E) Bar graph of EEF1A1 mRNA expression levels in asynchronously growing/cycling and serum-starved/G1-arrested MCF10A and MCF7 cells, as determined by qRT-PCR. Data are normalized to cycling MCF10A cells. P-values: Student t-test. *p < 0.05; ****p < 0.0001. (F) Graph showing oscillating, cell cycle stage-dependent mRNA expression levels of EEF1A1, the S-phase marker PCNA and the G2/M marker FBXO5. (G) Box plot of PCNA expression levels in breast carcinomas with indicated estrogen receptor (ER) status. Tumors were split into high and low EEF1A1 mRNA expression groups using median expression level as the cut-off. Data are derived from TCGA. P-values: Mann-Whitney U test. N/s, not significant; ****p < 0.0001. (H) Model for the relationship between estrogen receptor signaling and EEF1A1 mRNA expression in ER+ breast cancers.
Figure 4
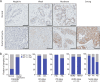
Immunohistochemistry shows that eEF1α1 protein is overexpressed in ductal breast carcinoma. (A) A tissue microarray with a total of 7 adjacent normal breast tissues and 100 ductal breast carcinomas was immunohistochemically stained with an anti-eEF1α1 antibody. Representative images of various staining intensities are shown. Scale bars: 50 μm. (B) Distributions of high and low eEF1α1-expressing tissue samples, using an immunohistochemistry-derived H-score of 50 as a cutoff. Data are analyzed for several clinical parameters, as indicated. P-values: Fisher’s exact tests or Chi-square tests, as indicated.
Figure 5
EEF1α1 protein expression levels parallel cellular stress levels. (A) Western blots showing eEF1α1 and β-actin protein levels in MCF10A and MDA-MB-231 breast cancer cell lines. (B) Quantification of β-actin-normalized eEF1α1 protein levels in MCF10A and MDA-MB-231 cell lines using the Western blots shown in (A). (C) Bar graph of normalized eEF1α1 protein levels in MCF10A and MDA-MB-231 cell lines using mass spectrometric quantification (each n = 2). (D) Protein expression levels of the heat shock response-inducible HSP90 isoforms α1 and α2. (E) Model explaining the contradictory EEF1A1 mRNA and eEF1α1 protein misexpression in breast carcinoma.
Similar articles
- Keratin 17 is overexpressed and predicts poor survival in estrogen receptor-negative/human epidermal growth factor receptor-2-negative breast cancer.
Merkin RD, Vanner EA, Romeiser JL, Shroyer ALW, Escobar-Hoyos LF, Li J, Powers RS, Burke S, Shroyer KR. Merkin RD, et al. Hum Pathol. 2017 Apr;62:23-32. doi: 10.1016/j.humpath.2016.10.006. Epub 2016 Nov 2. Hum Pathol. 2017. PMID: 27816721 - Id4 messenger RNA and estrogen receptor expression: inverse correlation in human normal breast epithelium and carcinoma.
de Candia P, Akram M, Benezra R, Brogi E. de Candia P, et al. Hum Pathol. 2006 Aug;37(8):1032-41. doi: 10.1016/j.humpath.2006.03.004. Epub 2006 May 22. Hum Pathol. 2006. PMID: 16867866 - Quantitative determination, by real-time reverse transcription polymerase chain reaction, of aromatase mRNA in invasive ductal carcinoma of the breast.
Zhang Z, Yamashita H, Toyama T, Omoto Y, Sugiura H, Hara Y, Wu X, Kobayashi S, Iwase H. Zhang Z, et al. Breast Cancer Res. 2003;5(6):R250-6. doi: 10.1186/bcr657. Epub 2003 Oct 9. Breast Cancer Res. 2003. PMID: 14580261 Free PMC article. - Prostate-specific membrane antigen expression in tumor-associated vasculature of breast cancers.
Wernicke AG, Varma S, Greenwood EA, Christos PJ, Chao KS, Liu H, Bander NH, Shin SJ. Wernicke AG, et al. APMIS. 2014 Jun;122(6):482-9. doi: 10.1111/apm.12195. Epub 2013 Dec 5. APMIS. 2014. PMID: 24304465
Cited by
- SKA3 Expression as a Prognostic Factor for Patients with Pancreatic Adenocarcinoma.
Buchholz K, Durślewicz J, Klimaszewska-Wiśniewska A, Wiśniewska M, Słupski M, Grzanka D. Buchholz K, et al. Int J Mol Sci. 2024 May 9;25(10):5134. doi: 10.3390/ijms25105134. Int J Mol Sci. 2024. PMID: 38791174 Free PMC article. - VGLL3 expression is associated with macrophage infiltration and predicts poor prognosis in epithelial ovarian cancer.
Haque R, Lee J, Chung JY, Shin HY, Kim H, Kim JH, Yun JW, Kang ES. Haque R, et al. Front Oncol. 2023 Jun 5;13:1152991. doi: 10.3389/fonc.2023.1152991. eCollection 2023. Front Oncol. 2023. PMID: 37342190 Free PMC article. - Investigating the Function of Human Jumping Translocation Breakpoint Protein (hJTB) and Its Interacting Partners through In-Solution Proteomics of MCF7 Cells.
Jayathirtha M, Whitham D, Alwine S, Donnelly M, Neagu AN, Darie CC. Jayathirtha M, et al. Molecules. 2022 Nov 28;27(23):8301. doi: 10.3390/molecules27238301. Molecules. 2022. PMID: 36500393 Free PMC article. - The progress of protein synthesis factors eIFs, eEFs and eRFs in inflammatory bowel disease and colorectal cancer pathogenesis.
Huang C, Zhao Q, Zhou X, Huang R, Duan Y, Haybaeck J, Yang Z. Huang C, et al. Front Oncol. 2022 Oct 31;12:898966. doi: 10.3389/fonc.2022.898966. eCollection 2022. Front Oncol. 2022. PMID: 36387239 Free PMC article. Review. - GBAS Regulates the Proliferation and Metastasis of Ovarian Cancer Cells by Combining with eEF1A1.
Ning X, Shi G, Ren S, Liu S, Ding J, Zhang R, Li L, Xie Q, Xu W, Meng F, Ma R. Ning X, et al. Oncologist. 2022 Feb 3;27(1):e64-e75. doi: 10.1093/oncolo/oyab015. Oncologist. 2022. PMID: 35305106 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous