Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations - PubMed (original) (raw)
doi: 10.1186/s13059-018-1522-1.
Bayazit Yunusbayev 2 3, Georgi Hudjashov 2 4, Anne-Mai Ilumäe 2, Siiri Rootsi 2, Terhi Honkola 5 6, Outi Vesakoski 5, Quentin Atkinson 7 8, Pontus Skoglund 9, Alena Kushniarevich 2 10, Sergey Litvinov 2 11, Maere Reidla 2 12, Ene Metspalu 2, Lehti Saag 2 12, Timo Rantanen 13, Monika Karmin 2, Jüri Parik 2 12, Sergey I Zhadanov 2 14, Marina Gubina 2 15, Larisa D Damba 2 16, Marina Bermisheva 2 11, Tuuli Reisberg 2, Khadizhat Dibirova 2 17, Irina Evseeva 18 19, Mari Nelis 20, Janis Klovins 21, Andres Metspalu 20, Tõnu Esko 20, Oleg Balanovsky 17 22, Elena Balanovska 17, Elza K Khusnutdinova 11 23, Ludmila P Osipova 15 24, Mikhail Voevoda 15 24 25, Richard Villems 2 12, Toomas Kivisild 2 12 26 27, Mait Metspalu 2
Affiliations
- PMID: 30241495
- PMCID: PMC6151024
- DOI: 10.1186/s13059-018-1522-1
Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations
Kristiina Tambets et al. Genome Biol. 2018.
Abstract
Background: The genetic origins of Uralic speakers from across a vast territory in the temperate zone of North Eurasia have remained elusive. Previous studies have shown contrasting proportions of Eastern and Western Eurasian ancestry in their mitochondrial and Y chromosomal gene pools. While the maternal lineages reflect by and large the geographic background of a given Uralic-speaking population, the frequency of Y chromosomes of Eastern Eurasian origin is distinctively high among European Uralic speakers. The autosomal variation of Uralic speakers, however, has not yet been studied comprehensively.
Results: Here, we present a genome-wide analysis of 15 Uralic-speaking populations which cover all main groups of the linguistic family. We show that contemporary Uralic speakers are genetically very similar to their local geographical neighbours. However, when studying relationships among geographically distant populations, we find that most of the Uralic speakers and some of their neighbours share a genetic component of possibly Siberian origin. Additionally, we show that most Uralic speakers share significantly more genomic segments identity-by-descent with each other than with geographically equidistant speakers of other languages. We find that correlated genome-wide genetic and lexical distances among Uralic speakers suggest co-dispersion of genes and languages. Yet, we do not find long-range genetic ties between Estonians and Hungarians with their linguistic sisters that would distinguish them from their non-Uralic-speaking neighbours.
Conclusions: We show that most Uralic speakers share a distinct ancestry component of likely Siberian origin, which suggests that the spread of Uralic languages involved at least some demic component.
Keywords: Genome-wide analysis; Haplotype analysis; IBD-segments; Population genetics; Uralic languages.
Conflict of interest statement
Ethics approval and consent to participate
DNA samples were obtained from unrelated volunteers, all donors provided informed consent and all experiments were performed in accordance with the relevant guidelines and regulations of the involved countries. The research has been approved by the Research Ethics Commitees of the University of Tartu and the Russian Academy of Sciences (approval nos. 228/M-40, 252/M-17, 17146-9217). Experimental methods of the study comply with the Helsinki Declaration.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
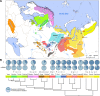
Geographic distribution of the Uralic-speaking populations and the schematic tree of the Uralic languages. a The geographic spread of the Uralic-speaking populations. Colour coding corresponds to the respective language in panel b. b Schematic representation of the phylogeny of the Uralic languages. Pie diagrams indicate the relative share of West and East Eurasian mitochondrial (mtDNA) and Y chromosomal (Y) lineages. Data from Additional file 5: Table S4 and Additional file 6: Table S5
Fig. 2
Principal component analysis (PCA) and genetic distances of Uralic-speaking populations. a PCA (PC1 vs PC2) of the Uralic-speaking populations (highlighted, population abbreviations are as in Additional file 1: Table S1). Values in brackets along the axes indicate the proportion of genetic variation explained by the components. b UPGMA tree of _F_ST distances calculated based on autosomal genetic variation
Fig. 3
Population structure of Uralic-speaking populations inferred from ADMIXTURE analysis on autosomal SNPs in Eurasian context. a Individual ancestry estimates for populations of interest for selected number of assumed ancestral populations (K3, K6, K9, K11). Ancestry components discussed in a main text (k2, k3, k5, k6, k9, k11) are indicated and have the same colours throughout. The names of the Uralic-speaking populations are indicated with blue (Finno-Ugric) or orange (Samoyedic). The full bar plot is presented in Additional file 3: Figure S3. b Frequency map of component k9
Fig. 4
Share of ~ 1–2 cM identity-by-descent (IBD) segments within and between regional groups of Uralic speakers. For each Uralic-speaking population representing lines in this matrix, we performed permutation test to estimate if it shows higher IBD segment sharing with other population (listed in columns) as compared to their geographic control group. Empty rectangles indicate no excess IBD sharing, rectangles filled in blue indicate comparisons when statistically significant excess IBD sharing was detected between one Uralic-speaking population with another Uralic-speaking population (listed in columns), rectangles filled in green mark the comparisons when a Uralic-speaking population shows excess IBD sharing with a non-Uralic-speaking population. For each tested Uralic speaker (matrix rows) populations in the control group that were used to generate permuted samples are indicated using small circles. For example, the rectangle filled in blue for Vepsians and Komis (A) implies that the Uralic-speaking Vepsians share more IBD segments with the Uralic-speaking Komis than the geographic control group for Vepsians, i.e. populations indicated with small circles (Central and North Russians, Swedes, Latvians and Lithuanians). The rectangle filled in green for Vepsians and Dolgans shows that the Uralic-speaking Vepsians share more IBD segments with the non-Uralic-speaking Dolgans than the geographic control group
Fig. 5
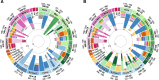
Circos plots of GLOBETROTTER (GT) results. The outer circle represents target groups for which GT inference was performed (wide segments) and additional surrogate populations, which were used to describe admixture in target populations (narrow segments). Geographic affiliation of target groups is colour-coded: blue—Europe (except populations from Volga-Ural region—Komis, Udmurts, Maris, Tatars, Chuvashes, Bashkirs); green—Volga-Ural region; and magenta—Western Siberia. Inner bar plots depict genetic composition of inferred sources of admixture in each of the target groups. A pair of sources is shown for a simple one-way admixture event between two populations, and an additional pair of sources for the less strongly signaled event is shown for a one-date multi-way admixture between more than two sources (marked as MW in the outer circle). In a simple one-date event, a pair of sources contributes 100% of the DNA of the target population. Surrogate populations in the inner bar plots are shaded according to the colour scheme given in the outer ring, and those contributing < 3% to mixing sources are coloured in grey. Point estimates and confidence intervals for the date of inferred admixture event are shown next to the cluster label. The details of the GT source groups are given in Additional file 3: Figure S5 and Additional file 11: Table S10. a Results of ‘full’ analysis, where each cluster was allowed to copy from every other cluster. b Results of ‘regional’ analysis, where no copying between samples from the same geographical region was allowed. For example, in the ‘full’ analysis of the ‘Europe 1’ cluster, a simple one-date admixture event was detected. The first source population contributes 85% of the total DNA, including 76% from the ‘Europe 2’ surrogate; the second source contributes 15% and is dominated by the ‘Finnic’ cluster. The admixture took place around 1211 CE (95% CI: 1213–1412 CE). Abbreviations: C-Central; Cauc-Caucasus; E-East; N-North; S-South; Sib-Siberia; W-West.
Fig. 6
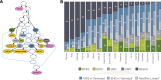
Proportions of ancestral components in studied European and Siberian populations and the tested qpGraph model. a The qpGraph model fitting the data for the tested populations. Colour codes for the terminal nodes: pink—modern populations (‘Population X’ refers to test population) and yellow—ancient populations (aDNA samples and their pools). Nodes coloured other than pink or yellow are hypothetical intermediate populations. We putatively named nodes which we used as admixture sources using the main recipient among known populations. The colours of intermediate nodes on the qpGraph model match those on the admixture proportions panel. b Admixture proportions (%) of ancestral components. We calculated the admixture proportions summing up the relative shares of a set of intermediate populations to explain the full spectrum of admixture components in the test population. We further did the same for the intermediate node CWC’ and present the proportions of the mixing three components in the stacked column bar of CWC’. Colour codes for ancestral components are as follows: dark green—Western hunter gatherer (WHG’); light green—Eastern hunter gatherer (EHG’); grey—European early farmer (LBK’); dark blue—carriers of Corded Ware culture (CWC’); and dark grey—Siberian. CWC’ consists of three sub-components: blue—Caucasian hunter-gatherer in Yamnaya (CHGinY’); light blue—Eastern hunter-gatherer in Yamnaya (EHGinY’); and light grey—Neolithic Levant (NeolL’)
Similar articles
- Between Lake Baikal and the Baltic Sea: genomic history of the gateway to Europe.
Triska P, Chekanov N, Stepanov V, Khusnutdinova EK, Kumar GPA, Akhmetova V, Babalyan K, Boulygina E, Kharkov V, Gubina M, Khidiyatova I, Khitrinskaya I, Khrameeva EE, Khusainova R, Konovalova N, Litvinov S, Marusin A, Mazur AM, Puzyrev V, Ivanoshchuk D, Spiridonova M, Teslyuk A, Tsygankova S, Triska M, Trofimova N, Vajda E, Balanovsky O, Baranova A, Skryabin K, Tatarinova TV, Prokhortchouk E. Triska P, et al. BMC Genet. 2017 Dec 28;18(Suppl 1):110. doi: 10.1186/s12863-017-0578-3. BMC Genet. 2017. PMID: 29297395 Free PMC article. - Y-chromosomal connection between Hungarians and geographically distant populations of the Ural Mountain region and West Siberia.
Post H, Németh E, Klima L, Flores R, Fehér T, Türk A, Székely G, Sahakyan H, Mondal M, Montinaro F, Karmin M, Saag L, Yunusbayev B, Khusnutdinova EK, Metspalu E, Villems R, Tambets K, Rootsi S. Post H, et al. Sci Rep. 2019 May 24;9(1):7786. doi: 10.1038/s41598-019-44272-6. Sci Rep. 2019. PMID: 31127140 Free PMC article. - More Rule than Exception: Parallel Evidence of Ancient Migrations in Grammars and Genomes of Finno-Ugric Speakers.
Santos P, Gonzàlez-Fortes G, Trucchi E, Ceolin A, Cordoni G, Guardiano C, Longobardi G, Barbujani G. Santos P, et al. Genes (Basel). 2020 Dec 11;11(12):1491. doi: 10.3390/genes11121491. Genes (Basel). 2020. PMID: 33322364 Free PMC article. - Integrating Linguistic, Archaeological and Genetic Perspectives Unfold the Origin of Ugrians.
Török T. Török T. Genes (Basel). 2023 Jun 26;14(7):1345. doi: 10.3390/genes14071345. Genes (Basel). 2023. PMID: 37510249 Free PMC article. Review. - The human genetic history of East Asia: weaving a complex tapestry.
Stoneking M, Delfin F. Stoneking M, et al. Curr Biol. 2010 Feb 23;20(4):R188-93. doi: 10.1016/j.cub.2009.11.052. Curr Biol. 2010. PMID: 20178766 Review.
Cited by
- Y chromosome sequencing data suggest dual paths of haplogroup N1a1 into Finland.
Preussner A, Leinonen J, Riikonen J, Pirinen M, Tukiainen T. Preussner A, et al. Eur J Hum Genet. 2024 Oct 28. doi: 10.1038/s41431-024-01707-7. Online ahead of print. Eur J Hum Genet. 2024. PMID: 39465313 - Population Characteristics of the Spectrum and Frequencies of CFTR Gene Mutations in Patients with Cystic Fibrosis from the Republic of Bashkortostan (Russia).
Ayupova G, Litvinov S, Akhmetova V, Minniakhmetov I, Mokrysheva N, Khusainova R. Ayupova G, et al. Genes (Basel). 2024 Oct 17;15(10):1335. doi: 10.3390/genes15101335. Genes (Basel). 2024. PMID: 39457459 Free PMC article. - Long shared haplotypes identify the Southern Urals as a primary source for the 10th century Hungarians.
Gyuris B, Vyazov L, Türk A, Flegontov P, Szeifert B, Langó P, Mende BG, Csáky V, Chizhevskiy AA, Gazimzyanov IR, Khokhlov AA, Kolonskikh AG, Matveeva NP, Ruslanova RR, Rykun MP, Sitdikov A, Volkova EV, Botalov SG, Bugrov DG, Grudochko IV, Komar O, Krasnoperov AA, Poshekhonova OE, Chikunova I, Sungatov F, Stashenkov DA, Zubov S, Zelenkov AS, Ringbauer H, Cheronet O, Pinhasi R, Akbari A, Rohland N, Mallick S, Reich D, Szécsényi-Nagy A. Gyuris B, et al. bioRxiv [Preprint]. 2024 Jul 23:2024.07.21.599526. doi: 10.1101/2024.07.21.599526. bioRxiv. 2024. PMID: 39091721 Free PMC article. Preprint. - Comparing Phylogeographies to Reveal Incompatible Geographical Histories within Genomes.
Singer B, Di Nardo A, Hein J, Ferretti L. Singer B, et al. Mol Biol Evol. 2024 Jul 3;41(7):msae126. doi: 10.1093/molbev/msae126. Mol Biol Evol. 2024. PMID: 38922185 Free PMC article. - Bronze age Northern Eurasian genetics in the context of development of metallurgy and Siberian ancestry.
Childebayeva A, Fricke F, Rohrlach AB, Huang L, Schiffels S, Vesakoski O, Mannermaa K, Semerau L, Aron F, Solodovnikov K, Rykun M, Moiseyev V, Khartanovich V, Kovtun I, Krause J, Kuzminykh S, Haak W. Childebayeva A, et al. Commun Biol. 2024 Jun 11;7(1):723. doi: 10.1038/s42003-024-06343-x. Commun Biol. 2024. PMID: 38862782 Free PMC article.
References
- Indreko R. Origin and area of settlement of the Fenno-Ugrian peoples. Science in Exile. Publication of the Scientific Quarterly “Scholar”. Heidelberg: Heidelberger Gutenberg-Druckerei GmbH; 1948. pp. 3–24.
- Setälä N. E (1926) Johdanto. In: Kannisto A, editor. Suomen suku I. Helsinki: Kustannusosakeyhtiö Otava.
Publication types
MeSH terms
Grants and funding
- PUT1217/Eesti Teadusagentuur/International
- PUT1339/Eesti Teadusagentuur/International
- IUT24/Eesti Teadusagentuur/International
- 2014-2020.4.01.15-0012/European Regional Development Fund/International
- 014-2020.4.01.16-0271/European Regional Development Fund/International
- 2014-2020.4.01.16-0125/European Regional Development Fund/International
- 0324-2018-0016/Russian Federation State Research Project/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- 16-06-00303/Russian Foundation for Basic Research/International
LinkOut - more resources
Full Text Sources
Other Literature Sources