ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING - PubMed (original) (raw)
. 2018 Nov 5;215(11):2868-2886.
doi: 10.1084/jem.20171029. Epub 2018 Sep 25.
Florian Tran 1 2, Go Ito 1 3, Raheleh Sheibani-Tezerji 1, Simone Lipinski 1, Jan W Kuiper 1, Markus Tschurtschenthaler 4 5, Svetlana Saveljeva 5, Joya Bhattacharyya 5, Robert Häsler 1, Kareen Bartsch 6, Anne Luzius 1, Marlene Jentzsch 1, Maren Falk-Paulsen 1, Stephanie T Stengel 1, Lina Welz 1, Robin Schwarzer 7, Björn Rabe 6, Winfried Barchet 8, Stefan Krautwald 9, Gunther Hartmann 8, Manolis Pasparakis 7, Richard S Blumberg 10, Stefan Schreiber 1 2, Arthur Kaser 5, Philip Rosenstiel 11
Affiliations
- PMID: 30254094
- PMCID: PMC6219748
- DOI: 10.1084/jem.20171029
ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING
Konrad Aden et al. J Exp Med. 2018.
Abstract
A coding variant of the inflammatory bowel disease (IBD) risk gene ATG16L1 has been associated with defective autophagy and deregulation of endoplasmic reticulum (ER) function. IL-22 is a barrier protective cytokine by inducing regeneration and antimicrobial responses in the intestinal mucosa. We show that ATG16L1 critically orchestrates IL-22 signaling in the intestinal epithelium. IL-22 stimulation physiologically leads to transient ER stress and subsequent activation of STING-dependent type I interferon (IFN-I) signaling, which is augmented in Atg16l1 ΔIEC intestinal organoids. IFN-I signals amplify epithelial TNF production downstream of IL-22 and contribute to necroptotic cell death. In vivo_,_ IL-22 treatment in Atg16l1 ΔIEC and Atg16l1 ΔIEC/Xbp1 ΔIEC mice potentiates endogenous ileal inflammation and causes widespread necroptotic epithelial cell death. Therapeutic blockade of IFN-I signaling ameliorates IL-22-induced ileal inflammation in Atg16l1 ΔIEC mice. Our data demonstrate an unexpected role of ATG16L1 in coordinating the outcome of IL-22 signaling in the intestinal epithelium.
© 2018 Aden et al.
Figures
Graphical abstract
Figure 1.
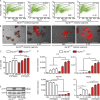
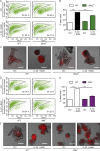
IL-22 induces cell death and a proinflammatory signature in Atg16l1-deficient intestinal organoids. (A) Representative FACS plots of PI-stained dissociated cells from intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC), treated with rmIL-22 (100 ng/ml) for 24 h. (B) Representative pictures of intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC), treated with rmIL-22 (100 ng/ml) for 24 h, stained with PI. Bars, 200 µm. (C) Flow cytometry assessment of dead cells from intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC) stimulated with rmIL-22 (1, 10, or 100 ng/ml) for 24 h using PI (n = 3 each). (D) mRNA expression of Tnf, Cxcl1, Atg16l1, and Reg3g in small intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC) treated with rmIL-22 (1, 10, or 100 ng/ml) for 24 h as assessed by qPCR (n = 4 each). (E) Western blot analysis from intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC) treated with rmIL-22 (100 ng/ml) for 30 min, probed against pSTAT3 and GAPDH. Illustrated are representative data of three independent experiments. Significance determined using Mann-Whitney test and expressed as the mean ± SEM. **, P < 0.01; ***, P < 0.001.
Figure 2.
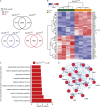
Atg16l1 orchestrates an IL-22–dependent IFN-I signature in intestinal organoids. (A) Venn diagram showing numbers of differentially expressed transcripts (overall, black) and significantly up-regulated (below, red) and down-regulated (below, blue) transcripts in small intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC) in response to stimulation with IL-22 (10 ng/ml) for 24 h (n = 4 each). Differential expression was determined using RNA sequencing and the DESEQ2 algorithm. (B) Heat map showing clustering of top 25 up- and down-regulated genes in response to IL-22 (10 ng/ml) according to genotype. (C) Gene set enrichment (GO) analysis of top 250 uniquely up-regulated genes in IL-22–treated _Atg16l1_ΔIEC intestinal organoids. (D) STRING-based network analysis of all genes contributing to the GO term “innate immune response” detected in C. Note a strong contribution of an IFN-I–related signature.
Figure 3.
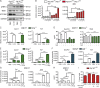
ATG16L1 coordinates an IL-22–dependent IFN-I signature via STING signaling. (A and B) Representative pictures (A) and quantification (B) of dsDNA in Caco-2 cells (ATG16L1+/+ vs. _ATG16L1_−/−) treated with rhIL-22 (100 ng/ml) for 24 h (n = 3 each). dsDNA was visualized in Caco-2 cells using an anti-dsDNA antibody (second antibody: Alexa Fluor 488–conjugated anti-mouse). Arrows indicate representative cytoplasmic dsDNA spots. Bars, 10 µm. (C) qPCR of Ifit1 and Ifit3 in intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC, _Sting_gt, _Mda5_−/−) treated with or without rmIL-22 (1, 10, or 100 ng/ml) for 24 h (n = 3 each). (D) qPCR of Ifit1 in intestinal organoids (C57BL/6, Cgas−/−, Irf3−/−, Il28r−/−) treated with rmIL-22 (100 ng/ml) or PBS for 24 h (n = 3 each). (E) qPCR of Sting in intestinal organoids (_Atg16l11_fl/fl, _Atg16l1_ΔIEC) treated with rmIL-22 (1, 10, or 100 ng/ml; n = 3 each). (F) Protein lysates from intestinal organoids (_Atg16l1_fl/fl, _Atg16l1_ΔIEC) treated with either rmIL-22 (100 ng/ml) or hydroxyurea (HU; 2 µM) for 24 h were subjected to immunoblot analysis against STING. (G) Western blot analyses from intestinal organoids (_Atg16l11_fl/fl, _Atg16l1_ΔIEC) treated with rmIL-22 (100 ng/ml) for 24 h. Lysates were probed against pTBK1, TBK1, ATG16L1, and GAPDH. (H) Protein lysates from Caco-2 cells (ATG16L1+/+ vs. _ATG16L1_−/−), treated with IL-22 (100 ng/ml) for indicated time points were subjected to immunoblot analysis against indicated proteins. LE: longer exposure. (I) qPCR of Ifit1 and Ifit3 in intestinal organoids (C57BL/6J) treated with rmIL-22 (100 ng/ml) and BafA (5 nM) for 24 h (n = 3 each). Results (A–I) represent at least two independent experiments. Significance determined using two-tailed Student’s t test and expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
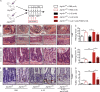
Figure 4.
IL-22 induces ileal inflammation in _Atg16l1_ΔIEC mice. (A) Stimulation scheme of _Atg16l1_fl/fl and _Atg16l1_ΔIEC mice (n = 5/5/5/5). Mice were treated with 2 µg/20 mg bodyweight of rmIL-22 i.p. every day over the course of 6 d. (B) Weight loss curve. (C) Statistical evaluation of the histological inflammation score in colon sections. (D–K) Histological evaluation of small intestinal sections with representative pictures and absolute quantification for H&E (D and E), TUNEL (F and G), and γH2AX (H and I; n = 5 each). Representative IF staining and statistical evaluation of small intestinal sections stained against pTBK1 (second antibody: Alexa Fluor 546–conjugated anti-rabbit; red) and counterstained with DAPI and anti-E-cadherin (second antibody: Alexa Fluor 488–conjugated anti-mouse; green; J and K; n = 5 each). For quantification, a minimum of 100 crypts/intestine were assessed in each treatment group by two independent observers. Bars, 100 µm. (L) Gene expression of Tnf, Ifit1, Ifit3, and Cxcl10 from small intestinal crypts (n = 4 each). Results represent one experiment. Significance determined using two-tailed Student’s t test and expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 5.
IL-22 aggravates epithelial cell death–mediated inflammation in _Atg16l1_ΔIEC/_Xbp1_ΔIEC mice. (A) Treatment scheme of _Atg16l1_fl/fl/_Xbp1_fl/fl and Atg161_ΔIEC/Xbp1_ΔIEC mice (n = 7/7/7/6). (B–K) Histological evaluation of small intestinal sections with representative pictures and absolute quantification for H&E (B and C), TUNEL (D and E), and γH2AX (F and G; n = 5 each). Representative IF staining (including magnification inserts) and statistical evaluation of small intestinal sections stained against dsDNA (second antibody: Alexa Fluor 488–conjugated anti-mouse antibody; green; counterstained with DAPI; H and I) and pTBK1 (second antibody: Alexa Fluor 546–conjugated anti-rabbit; red), counterstained with DAPI and anti–E-cadherin (second antibody: Alexa Fluor 488–conjugated anti-mouse; green; J and K; n = 5 each). For quantification a minimum of 100 crypts/intestine were assessed in each treatment group. Bars, 100 µm. (L) qPCR of Ifnb and Tnf in ileal mucosa (n = 4 each). Results represent one experiment. Significance determined using two-tailed Student’s t test and expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 6.
STING and IFN-I signals synergize in TNF induction and necroptosis in intestinal epithelial organoids. (A) Immunoblot analyses from protein lysates derived from _Atg16l1_fl/fl/_Xbp1_fl/fl and _Atg16l1_ΔIEC/_Xbp1_ΔIEC organoids stimulated with rmIL-22 (100 ng/ml) for 24 h and probed against pTBK1, TBK1, ATG16L1, and GAPDH. (B) Transcript levels of Tnf and Mlkl in small intestinal organoids (_Atg16l1_fl/fl/_Xbp1_fl/fl and _Atg16l1_ΔIEC/_Xbp1_ΔIEC) treated with rmIL-22 (100 ng/ml) for 24 h as assessed by qPCR (n = 4 each). (C) Transcript levels of Cxcl1, Tnf in small intestinal organoids (_C57BL/6, Sting_gt) treated with rmIL-22 (100 ng/ml) for 24 h as assessed by qPCR (n = 4 each). (D) Concentration of CXCL10 and TNF in the supernatant of intestinal organoids (_C57BL/6, Sting_gt) treated with rmIL-22 (100 ng/ml), bafilomycin A (BafA; 5 nM) or both for 24 h, as detected via ELISA (n = 3 each). (E) qPCR of Cxcl1, Tnf in small intestinal organoids (_Sting_gt) treated with rmIL-22 (100 ng/ml) or IFN-β (1,000 IU/ml) or both for 24 h (n = 3 each). (F) qPCR of Cxcl10, Tnf in small intestinal organoids from C57BL/6 or Il28r−/− mice treated with rmIL-22 (100 ng/ml) for 24 h (n = 3 each). (G) qPCR of Mlkl and Tnf in small intestinal organoids (C57BL/6) treated with rmIL-22 (100 ng/ml) or IFN-β (1,000 IU/ml) or both for 24 h (n = 3 each). (H) Concentration of TNF in the supernatant of intestinal organoids (C57BL/6) treated with rmIL-22 (100 ng/ml) or IFN-β (1,000 IU/ml) or both for 24 h, as detected via ELISA. (I) Assessment of dead cells from intestinal organoids (_Atg16l1_ΔIEC) stimulated with rmIL-22 (100 ng/ml) for 24 h in the absence or presence of anti-TNF antibody (10 and 100 ng/ml; n = 3 each). Results represent two independent experiments. Significance determined using two-tailed Student’s t test and expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 7.
IL-22–induced epithelial cell death depends on STING and MLKL. (A and B) Representative FACS plots (A) and flow cytometry analysis (B) of cell death of intestinal organoids (WT, _Sting_gt), treated with rmIL-22 (100 ng/ml) and BafA (5 nM) for 24 h and staind with PI (n = 3 each). (C) Representative pictures of intestinal organoids (WT, _Sting_gt), treated with rmIL-22 (100 ng/ml) and BafA (5 nM) for 24 h. Bars, 200 µm. (D and E) Representative FACS plots (D) and flow cytometry analysis (E) of cell death of intestinal organoids (WT, _Mlkl_−/−), treated with rmIL-22 (100 ng/ml) and BafA (5 nM) for 24 h and stained with PI (n = 3 each). (F) Representative pictures of intestinal organoids (WT, _Mlkl_−/−), treated with rmIL-22 (100 ng/ml) and BafA (5 nM) for 24 h. Bars, 200 µm. Results represent two independent experiments. Significance determined using two-tailed Student’s t test and expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 8.
IFN-I signals contribute to IL-22–induced ileitis in Atg16l1ΔIEC mice. (A) Stimulation scheme of _Atg16l1_fl/fl and _Atg16l1_ΔIEC mice treated with rmIL-22 and anti-IFNAR. Mice were treated with either rmIL-22 i.p. (2 µg/20 mg bodyweight) or PBS on days 0, 2, 4, 6, and 8. A group of mice received anti-IFNAR i.p. (10 mg/kg bodyweight). All mice were terminated at day 10. (B and C) Histological evaluation of colonic section with representative pictures (B) and absolute quantification for H&E (C; n = 5/8/6/8/8). Bars, 500 µm (upper); 200 µm (lower). (D–G) Histological evaluation of small intestinal sections with representative pictures and absolute quantification for H&E (D and E) and TUNEL (F and G); n = 5/8/6/8/8). Bars, 100 µm. Results represent one experiment. Significance determined using two-tailed Student’s t test (C, E, and G) and expressed as the mean ± SEM. *, P < 0.05; ***, P < 0.001.
Figure 9.
The IL-22–IFN-I axis affects clinical outcome upon anti-TNF therapy in IBD patients. (A) Linear regression of correlation of IL22, TNF, and MLKL or a composite score (Lübbers et al., 2013) of six IFN stimulatory genes (ISG) in sigmoid biopsies from IBD patients (n = 21). (B) Relative mRNA expression of IL22, TNF, MLKL, or a composite score of six ISG in sigmoid biopsies from human IBD patients before (week 0) or after anti-TNF therapy (week 14) clustered according to clinical remission status (remission: UC = 7, CD = 7; nonremission: CD = 1, UC = 6). Significance determined using Spearman test for correlation (A) or Mann-Whitney test (B) and expressed as the mean ± SEM.
References
- Aden K., Rehman A., Falk-Paulsen M., Secher T., Kuiper J., Tran F., Pfeuffer S., Sheibani-Tezerji R., Breuer A., Luzius A., et al. 2016. Epithelial IL-23R Signaling Licenses Protective IL-22 Responses in Intestinal Inflammation. Cell Reports. 16:2208–2218. 10.1016/j.celrep.2016.07.054 - DOI - PMC - PubMed
- Bachmann M., Horn K., Rudloff I., Goren I., Holdener M., Christen U., Darsow N., Hunfeld K.-P., Koehl U., Kind P., et al. 2010. Early production of IL-22 but not IL-17 by peripheral blood mononuclear cells exposed to live Borrelia burgdorferi: the role of monocytes and interleukin-1. PLoS Pathog. 6:e1001144 10.1371/journal.ppat.1001144 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 DK044319/DK/NIDDK NIH HHS/United States
- R01 DK088199/DK/NIDDK NIH HHS/United States
- R01 DK051362/DK/NIDDK NIH HHS/United States
- R01 DK053056/DK/NIDDK NIH HHS/United States
- 106260/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- P30 DK034854/DK/NIDDK NIH HHS/United States
- Wellcome Trust/United Kingdom
- R01 DK044319/DK/NIDDK NIH HHS/United States
- R56 DK053056/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials