Jumbo Bacteriophages Are Represented Within an Increasing Diversity of Environmental Viruses Infecting the Emerging Phytopathogen, Dickeya solani - PubMed (original) (raw)
Jumbo Bacteriophages Are Represented Within an Increasing Diversity of Environmental Viruses Infecting the Emerging Phytopathogen, Dickeya solani
Andrew Day et al. Front Microbiol. 2018.
Abstract
Dickeya species are economically important phytopathogens widespread in mainland Europe that can reduce crop yields by 25%. There are no effective environmentally-acceptable chemical systems available for diseases caused by Dickeya. Bacteriophages have been suggested for use in biocontrol of these pathogens in the field, and limited field trials have been conducted. To date the majority of bacteriophages capable of infecting Dickeya solani, one of the more aggressive species, are from the same family, the Ackermannviridae, many representatives of which have been shown to be unsuitable for use in the field due to their capacity for generalized transduction. Members of this family are also only capable of forming individual plaques on D. solani. Here we describe novel bacteriophages from environmental sources isolated on D. solani, including members of two other viral families; Myoviridae and Podoviridae, most of which are capable of forming plaques on multiple Dickeya species. Full genomic sequencing revealed that the Myoviridae family members form two novel clusters of jumbo bacteriophages with genomes over 250 kbp, with one cluster containing phages of another phytopathogen Erwinia amylovora. Transduction experiments showed that the majority of the new environmental bacteriophages are also capable of facilitating efficient horizontal gene transfer, however the single Podoviridae family member is not. This particular phage therefore has potential for use as a biocontrol agent against multiple species of Dickeya.
Keywords: Ackermannviridae; Dickeya solani; bacteriophage; environmental viruses; horizontal gene transfer; jumbo bacteriophage; phage therapy; phytopathogen.
Figures
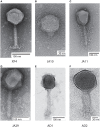
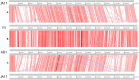
Figure 1
Transmission electron micrographs of six Dickeya solani phages. XF4 (A) is a member of the Ackermannviridae family, exhibiting an icosahedral head of around 90 nm, a tail length of 110 nm and tail spikes at the base of the tail. JA10 (B) is a member of the Podoviridae family, characterized by a short stubby tail. JA11, JA29, and AD1 (C–E) are members of the Myoviridae family with unclear tail appendages. AD2 (F) has a similar morphology to XF4 and has a partially-contracted tail.
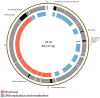
Figure 2
Map of the genome of JA10. The outer gray ring marks open reading frames, with those highlighted in black discussed in more detail in the text, whilst the inner ring categorizes the proposed functions of these genes. The genome map was generated using Circos.
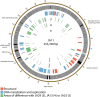
Figure 3
Map of the genome of JA11. The outer gray ring marks open reading frames, with those highlighted in black discussed in more detail in the text. The second ring categorizes the proposed functions of these genes, whilst the inner rings highlight the areas of the genome that differ from the genomes of JA29, JA13, and JA33 (third to fifth ring, respectively). The genome map was generated using Circos.
Figure 4
Translated nucleotide comparison of a putative DNA helicase between JA11 (top), JA33 (second), JA13 (third), and JA29 (bottom). Red bars mark areas of amino acid identity, with darker colors showing higher identity. Figure produced using the Artemis Comparison Tool.
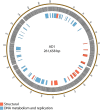
Figure 5
Map of the genome of AD1. The outer gray ring marks open reading frames whilst the inner ring categorizes the proposed functions of these genes. The genome map was generated using Circos.
Figure 6
Translated nucleotide comparison of the genomes of JA11 (A), Y3 (B), and AD1 (C). Red bars mark areas of amino acid identity, with darker colors showing higher identity. Blue bars highlight areas of inversion. Figure produced using the Artemis Comparison Tool.
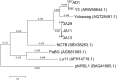
Figure 7
Phylogenetic tree of the large terminase subunit constructed using the Maximum Likelihood method in MEGA. All positions containing gaps and missing data were eliminated, with 642 positions in the final dataset. The tree shown has the highest log likelihood (−8677.52) and is drawn to scale, with branch lengths measured in the number of substitutions per site.
Figure 8
Phylogenetic tree of three tail fiber proteins constructed using the Maximum Likelihood method in MEGA. All positions containing gaps and missing data were eliminated, with 158 positions in the final dataset. The tree shown has the highest log likelihood (−2426.84) and is drawn to scale, with branch lengths measured in the number of substitutions per site.
Similar articles
- Environmental Bacteriophages of the Emerging Enterobacterial Phytopathogen, Dickeya solani, Show Genomic Conservation and Capacity for Horizontal Gene Transfer between Their Bacterial Hosts.
Day A, Ahn J, Fang X, Salmond GPC. Day A, et al. Front Microbiol. 2017 Aug 30;8:1654. doi: 10.3389/fmicb.2017.01654. eCollection 2017. Front Microbiol. 2017. PMID: 28912766 Free PMC article. - Newly Isolated Bacteriophages from the Podoviridae, Siphoviridae, and Myoviridae Families Have Variable Effects on Putative Novel Dickeya spp.
Alič Š, Naglič T, Tušek-Žnidarič M, Ravnikar M, Rački N, Peterka M, Dreo T. Alič Š, et al. Front Microbiol. 2017 Sep 28;8:1870. doi: 10.3389/fmicb.2017.01870. eCollection 2017. Front Microbiol. 2017. PMID: 29033917 Free PMC article. - Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani.
Carstens AB, Djurhuus AM, Kot W, Jacobs-Sera D, Hatfull GF, Hansen LH. Carstens AB, et al. Viruses. 2018 Nov 10;10(11):621. doi: 10.3390/v10110621. Viruses. 2018. PMID: 30423804 Free PMC article. - Intracellular Organization by Jumbo Bacteriophages.
Guan J, Bondy-Denomy J. Guan J, et al. J Bacteriol. 2020 Dec 18;203(2):e00362-20. doi: 10.1128/JB.00362-20. Print 2020 Dec 18. J Bacteriol. 2020. PMID: 32868402 Free PMC article. Review. - Bacteriophages of Soft Rot Enterobacteriaceae-a minireview.
Czajkowski R. Czajkowski R. FEMS Microbiol Lett. 2016 Jan;363(2):fnv230. doi: 10.1093/femsle/fnv230. Epub 2015 Nov 30. FEMS Microbiol Lett. 2016. PMID: 26626879 Review.
Cited by
- Emerging Aspects of Jumbo Bacteriophages.
Nazir A, Ali A, Qing H, Tong Y. Nazir A, et al. Infect Drug Resist. 2021 Nov 30;14:5041-5055. doi: 10.2147/IDR.S330560. eCollection 2021. Infect Drug Resist. 2021. PMID: 34876823 Free PMC article. Review. - The branched receptor-binding complex of Ackermannviridae phages promotes adaptive host recognition.
Sørensen AN, Woudstra C, Kalmar D, Poppeliers J, Lavigne R, Sørensen MCH, Brøndsted L. Sørensen AN, et al. iScience. 2024 Sep 7;27(9):110813. doi: 10.1016/j.isci.2024.110813. eCollection 2024 Sep 20. iScience. 2024. PMID: 39310758 Free PMC article. - fENko-Kae01 is a flagellum-specific jumbo phage infecting Klebsiella aerogenes.
Ranta K, Skurnik M, Kiljunen S. Ranta K, et al. BMC Microbiol. 2024 Jul 1;24(1):234. doi: 10.1186/s12866-024-03387-1. BMC Microbiol. 2024. PMID: 38951769 Free PMC article. - Isolation and Characterization of a Novel Jumbo Phage from Leaf Litter Compost and Its Suppressive Effect on Rice Seedling Rot Diseases.
Sasaki R, Miyashita S, Ando S, Ito K, Fukuhara T, Takahashi H. Sasaki R, et al. Viruses. 2021 Mar 31;13(4):591. doi: 10.3390/v13040591. Viruses. 2021. PMID: 33807245 Free PMC article. - Jumbo Phages: A Comparative Genomic Overview of Core Functions and Adaptions for Biological Conflicts.
M Iyer L, Anantharaman V, Krishnan A, Burroughs AM, Aravind L. M Iyer L, et al. Viruses. 2021 Jan 5;13(1):63. doi: 10.3390/v13010063. Viruses. 2021. PMID: 33466489 Free PMC article.
References
- Ackermann H.-W. (2012). Bacteriophage electron microscopy, in Advances in Virus Research, eds Lobocka M., Szybalski W. T. (Elsevier; ), 1–32. - PubMed
- Adeolu M., Alnajar S., Naushad S., Gupta R. S. (2016). Genome-based phylogeny and taxonomy of the “Enterobacteriales:” proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morgane. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. 10.1099/ijsem.0.001485 - DOI - PubMed
Grants and funding
- BB/G000298/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H002677/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources