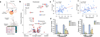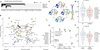Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy - PubMed (original) (raw)
. 2018 Oct;50(10):1399-1411.
doi: 10.1038/s41588-018-0209-6. Epub 2018 Sep 27.
Jin-Ku Lee 1 2 3, Jason K Sa 1 3, Sang Shin 1 6, Jiguang Wang 7 8 9, Mykola Bordyuh 4 5, Hee Jin Cho 1 3, Oliver Elliott 4 5, Timothy Chu 4 5, Seung Won Choi 1 6, Daniel I S Rosenbloom 4 5, In-Hee Lee 1 3, Yong Jae Shin 1 2 3, Hyun Ju Kang 1 3, Donggeon Kim 1 3, Sun Young Kim 10, Moon-Hee Sim 10, Jusun Kim 10, Taehyang Lee 10, Yun Jee Seo 1 3, Hyemi Shin 1 6, Mijeong Lee 1 6, Sung Heon Kim 1 2, Yong-Jun Kwon 1, Jeong-Woo Oh 1 6, Minsuk Song 1, Misuk Kim 1 3, Doo-Sik Kong 2, Jung Won Choi 2, Ho Jun Seol 2, Jung-Il Lee 2, Seung Tae Kim 10, Joon Oh Park 6 10, Kyoung-Mee Kim 11, Sang-Yong Song 11, Jeong-Won Lee 12, Hee-Cheol Kim 13, Jeong Eon Lee 13, Min Gew Choi 13, Sung Wook Seo 14, Young Mog Shim 15, Jae Ill Zo 15, Byong Chang Jeong 16, Yeup Yoon 3 6, Gyu Ha Ryu 3, Nayoung K D Kim 3 17, Joon Seol Bae 3 17, Woong-Yang Park 3 6 17, Jeongwu Lee 18, Roel G W Verhaak 19, Antonio Iavarone 20 21 22, Jeeyun Lee 23 24, Raul Rabadan 25 26, Do-Hyun Nam 27 28 29
Affiliations
- PMID: 30262818
- PMCID: PMC8514738
- DOI: 10.1038/s41588-018-0209-6
Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy
Jin-Ku Lee et al. Nat Genet. 2018 Oct.
Abstract
Outcomes of anticancer therapy vary dramatically among patients due to diverse genetic and molecular backgrounds, highlighting extensive intertumoral heterogeneity. The fundamental tenet of precision oncology defines molecular characterization of tumors to guide optimal patient-tailored therapy. Towards this goal, we have established a compilation of pharmacological landscapes of 462 patient-derived tumor cells (PDCs) across 14 cancer types, together with genomic and transcriptomic profiling in 385 of these tumors. Compared with the traditional long-term cultured cancer cell line models, PDCs recapitulate the molecular properties and biology of the diseases more precisely. Here, we provide insights into dynamic pharmacogenomic associations, including molecular determinants that elicit therapeutic resistance to EGFR inhibitors, and the potential repurposing of ibrutinib (currently used in hematological malignancies) for EGFR-specific therapy in gliomas. Lastly, we present a potential implementation of PDC-derived drug sensitivities for the prediction of clinical response to targeted therapeutics using retrospective clinical studies.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
The authors declare no competing financial interests.
Figures
Figure 1.. Patient tumor and derived cell resources for pharmacogenomics analysis.
(a) Overview of the procedure for pharmacogenomics analysis in patient tumor-derived cells (PDCs). A total of 462 PDCs from 14 cancer types were isolated. Genomic contexts were analyzed to identify somatic variants and/or gene expression profiles. Comprehensive genomic profiling from 14 cancer lineages are summarized in a circos plot, demonstrating detailed data structure and size for each type of available molecular data. Short-term cultured PDCs have undergone drug sensitivity screening to 60 molecular targeted agents. Clinical feasibility of PDC screening-guided precision therapy was evaluated. (b) Three-dimensional bubble plot showing the frequency of somatic nonsynonymous mutations exclusively in tissue (red; left axis), exclusively in PDC (black; right axis), and in common to the two (yellow; upper axis). 122 samples with DNA sequencing data from tumor tissue and PDC samples were considered in this analysis. (c) Comparison of mRNA expression profiles between primary tissue and PDC on 24 paired samples with matched RNASeq data. Spearman correlations of mRNA expression between tissue and PDC are shown as a heat map. Paired samples are located along the diagonal.
Figure 2.. Therapeutic landscape of PDCs and lineage-specific responses
(a) Tumor lineage-specific drug association identified using 60 compounds (n=462 biologically independent samples). Two-sided wilcox rank sum test was applied to determine the relative differences of drug sensitivity between specific tumor type and the rest. Only significant associations are marked (q-value < 0.05). Drugs are ordered based on their known targets. (b) A volcano plot representation of TDA analysis showing the magnitude (Fold change, x-axis) and significance (TDA q value, y-axis) of all tumor-drug associations (n=462 biologically independent samples). Each circle represents a single tumor-drug interaction and the size is proportional to the cohort size of that tumor. (c) Distribution of gastric, glioma PDCs and BYL719 drug AUC profile over the topological representation of PDCs (n=462 biologically independent samples). Each node represents a set of PDCs with similar AUC profiles. A PDC can appear in several nodes, and two nodes are connected by an edge if they have at least one PDC in common. The P values were calculated using the pearson correlation test between the fraction distribution of gastric or glioma cell lines and mean AUC values of BYL719 drug over the nodes and they were adjusted using BH method. (d) Violin plots measures the activity level of PI3K–AKT–mTOR pathway on gastric and GBMs using TCGA RNASeq datasets (n=100 biologically independent samples for each group). We adopt the enrichment score derived from ssGSEA analysis as assessment. The P value is calculated from two-sided wilcox rank-sum test. Horizontal lines within the violin plot represent 0.25, 0.50, and 0.75 quantiles.
Figure 3.. Pharmacogenomic interactions in PDCs.
(a) A volcano plot representation of correlation analysis showing the magnitude and significance of gene-drug associations (n=360 biologically independent samples). (b) Waterfall plot enumerating significant associations between KRAS mutation and drug sensitivity (n=360 biologically independent samples). Two horizontal dashed lines indicate statistical significance. (c) SW480 was treated with DMSO (control) or trametinib (0.1 or 1 mM), followed by incubation with two EGFR inhibitors, dacomitinib (left) and gefitinib (right). Cell viability for each dose was normalized to DMSO or trametinib (0.1 or 1 mM) treatment only cells. (d) Probability distribution of drug-target families over the topological network. Each node represents a set of drugs with similar AUC profiles. A drug can appear in several nodes, and two nodes are connected by an edge if they have at least one drug in common. Colors of the nodes correspond to mean RGB values of drug families. Ibrutinib belongs to three nodes on the network encompassed with an oval. (e) Drug sensitivities to ibrutinib in 67 PDCs. The red color in the heat map represents sensitivity, while the blue color indicates resistance. EGFR alterations including genomic amplification, vIII, and expression are shown. (f) Kaplan-Meier survival plots for P2.T (EGFR amp/vIII) orthotopic mice model. Once intracranial model was established, Vehicle (0.5% methylcellulose) or iburitnib (50mg/kg/day) was administrated orally (PO) for 5 consecutive days and 2 days of resting period per each cycle (n=8 per group). _P_-values: a,b,e, two-sided wilcox rank sum test. _P_-values: f, two-sided Log-rank test.
Figure 4.. Genomic and transcriptomic correlates of panobinostat sensitivity
(a) Schematic overview for dNetFS (Diffusion kernel based Network method for Feature Selection of drug sensitivity). In brief, dNetFS integrates genomic/pharmaceutical data, protein-protein interaction network, and prior knowledge of drug-targets interaction to prioritize genetic and gene expression features of PDCs that predict drug response. (b) Predictive features of panobinostat response identified by the dNetFS are plotted for their frequency and effect size. Associations are colored in red for expression features and blue for others. Node size is proportional to the single drug–feature linear correlation. (c) Scatter plot showing linear correlation between panobinostat AUC and HDAC4 expression (left panel) and SIK2 expression (right panel) (n=69 biologically independent samples). The correlation coefficient and the P values were obtained using pearson correlation test. (d) Drug response assessment of panobinostat (1.2nM or 4.8nM) with shRNA-mediated knockdown of HDAC4 or NT (non-target) (left panel) and SIK2 or NT (right panel). Cell viability for each dose was normalized to sole transduced cells only. Data are mean ± s.d. of n=3 technical replicates. Experiments were repeated three times with similar results. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, two-tailed t-test.
Figure 5.. Predictive biomarkers for response to EGFR inhibitors in EGFR altered GBM PDCs.
(a) Mutational landscape of EGFR alterations in GBM cohort. (b) For the 10 EGFR inhibitors, top drug–feature associations identified by dNetFS are plotted for their frequency and effect size (n=49 biologically independent samples in (a). Node size is proportional to the single drug–feature linear correlation. (c) Gene expression profiles of EGFR and NRG1, and AUC drug response profiles of erlotinib and dacomitinib, over the topological representation (n=44 biologically independent samples). (d) Drug response assessment of EGFR inhibitors with shRNA-mediated knockdown of NRG1 or NT (non-target). Cell viability for each dose was normalized to shNRG1 or NT transduced cells only. (n=10 independent experiments with 3 technical replicates) (e) Comparisons of cancer pathway activities between two _EGFR_-altered GBM subgroups that were most sensitive and most resistance. We adopt the enrichment score derived from ssGSEA analysis as assessment. (f) Drug response assessment of EGFR inhibitors with PI3K-AKT-mTOR (PAM) inhibitors or DMSO. Cell viability for each dose was normalized to PAM or DMSO treated cells only. Mean AUC value for 4 PAM inhibitors (BYL719, BKM120, BEZ235, and AZD2014) was plotted (n=8 independent experiments with 3 technical replicates). _P_-values: c, pearson correlation test. _P_-values: d,f, two-sided wilcox rank sum test. Horizontal lines within the violin plot represent 0.25, 0.50, and 0.75 quantiles.
Figure 6.. Clinical feasibility of PDC drug screening-guided precision oncology.
(a) Bar graph represents normalized AUCs (Z-scores from pan-cancer dataset) of indicated drugs (n=31 biologically independent samples). Clinical responses were determined according to RECIST. Multiple-target drugs are classified based on corresponding representative genomic targets. (b) Representative box plot of (a). CR (complete response); PR (partial response); SD (stable disease); PD (progressive disease) per RECIST criteria. Box plot spans from the first to third quartiles and the whiskers represent the 1.5 interquartile range. (c) Receiver operating characteristic (ROC) curve was plotted by the sensitivity (%) and 100-specificity (%) values for predicting clinical response rate using z-scores in (a). (d-f) T1-weighted contrast enhanced (T1CE), T2-flare magnetic resonance images (MRI) or computed tomography (CT) images for indicated patients before and after drug treatment were demonstrated. Circles on the bar represent days obtaining presented images, where 0D refers the day starting drug treatment. Negative (−) or positive (+) D represents days before or after treatment, respectively. Red arrows indicate measurable or progressed tumors, and orange arrows represent partial response. Vertical scattered plot for AUCs of indicated drugs in pan-cancer AUC reference dataset for the indicated drug. AUCs of PDCs isolated from the illustrated patients were highlighted. _P_-value: b, two-sided wilcox rank sum test. _P_-values: c, two-sided binomial exact test.
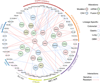
Figure 7.. Schematic illustration of major lineage-specific and genomic associated drug interactions.
Molecular targeting agents are clustered based on drug family classification and connected to various genomic alterations including mutation, mRNA expression, copy number variation (CNV), and fusion. Edges represent sensitive or resistant gene-drug interactions. Lineage-specific drug associations are highlighted in each drug node based on specific lineage type.
Comment in
- Cancer cell lines as patient avatars for drug response prediction.
McDermott U. McDermott U. Nat Genet. 2018 Oct;50(10):1350-1351. doi: 10.1038/s41588-018-0245-2. Nat Genet. 2018. PMID: 30262817 No abstract available. - An avatar for precision cancer therapy.
Kato S, Kurzrock R. Kato S, et al. Nat Biotechnol. 2018 Nov 9;36(11):1053-1055. doi: 10.1038/nbt.4293. Nat Biotechnol. 2018. PMID: 30412185 No abstract available.
Similar articles
- Patient-Derived Organoids from Multiple Colorectal Cancer Liver Metastases Reveal Moderate Intra-patient Pharmacotranscriptomic Heterogeneity.
Bruun J, Kryeziu K, Eide PW, Moosavi SH, Eilertsen IA, Langerud J, Røsok B, Totland MZ, Brunsell TH, Pellinen T, Saarela J, Bergsland CH, Palmer HG, Brudvik KW, Guren T, Dienstmann R, Guren MG, Nesbakken A, Bjørnbeth BA, Sveen A, Lothe RA. Bruun J, et al. Clin Cancer Res. 2020 Aug 1;26(15):4107-4119. doi: 10.1158/1078-0432.CCR-19-3637. Epub 2020 Apr 16. Clin Cancer Res. 2020. PMID: 32299813 - Patient-derived cell models as preclinical tools for genome-directed targeted therapy.
Lee JY, Kim SY, Park C, Kim NK, Jang J, Park K, Yi JH, Hong M, Ahn T, Rath O, Schueler J, Kim ST, Do IG, Lee S, Park SH, Ji YI, Kim D, Park JO, Park YS, Kang WK, Kim KM, Park WY, Lim HY, Lee J. Lee JY, et al. Oncotarget. 2015 Sep 22;6(28):25619-30. doi: 10.18632/oncotarget.4627. Oncotarget. 2015. PMID: 26296973 Free PMC article. - Precision Medicine in Pediatric Oncology.
Vo KT, Parsons DW, Seibel NL. Vo KT, et al. Surg Oncol Clin N Am. 2020 Jan;29(1):63-72. doi: 10.1016/j.soc.2019.08.005. Epub 2019 Oct 29. Surg Oncol Clin N Am. 2020. PMID: 31757314 Free PMC article. Review. - New tools for old drugs: Functional genetic screens to optimize current chemotherapy.
Gerhards NM, Rottenberg S. Gerhards NM, et al. Drug Resist Updat. 2018 Jan;36:30-46. doi: 10.1016/j.drup.2018.01.001. Epub 2018 Jan 12. Drug Resist Updat. 2018. PMID: 29499836 Free PMC article. Review. - Molecular Tumor Boards in Clinical Practice.
Luchini C, Lawlor RT, Milella M, Scarpa A. Luchini C, et al. Trends Cancer. 2020 Sep;6(9):738-744. doi: 10.1016/j.trecan.2020.05.008. Epub 2020 Jun 6. Trends Cancer. 2020. PMID: 32517959
Cited by
- High-Throughput Drug Screening of Primary Tumor Cells Identifies Therapeutic Strategies for Treating Children with High-Risk Cancer.
Mayoh C, Mao J, Xie J, Tax G, Chow SO, Cadiz R, Pazaky K, Barahona P, Ajuyah P, Trebilcock P, Malquori A, Gunther K, Avila A, Yun DY, Alfred S, Gopalakrishnan A, Kamili A, Wong M, Cowley MJ, Jessop S, Lau LMS, Trahair TN, Ziegler DS, Fletcher JI, Gifford AJ, Tsoli M, Marshall GM, Haber M, Tyrrell V, Failes TW, Arndt GM, Lock RB, Ekert PG, Dolman MEM. Mayoh C, et al. Cancer Res. 2023 Aug 15;83(16):2716-2732. doi: 10.1158/0008-5472.CAN-22-3702. Cancer Res. 2023. PMID: 37523146 Free PMC article. - Comprehensive pharmacogenomic characterization of gastric cancer.
Sa JK, Hong JY, Lee IK, Kim JS, Sim MH, Kim HJ, An JY, Sohn TS, Lee JH, Bae JM, Kim S, Kim KM, Kim ST, Park SH, Park JO, Lim HY, Kang WK, Her NG, Lee Y, Cho HJ, Shin YJ, Kim M, Koo H, Kim M, Seo YJ, Kim JY, Choi MG, Nam DH, Lee J. Sa JK, et al. Genome Med. 2020 Feb 18;12(1):17. doi: 10.1186/s13073-020-0717-8. Genome Med. 2020. PMID: 32070411 Free PMC article. - Integrated pharmaco-proteogenomics defines two subgroups in isocitrate dehydrogenase wild-type glioblastoma with prognostic and therapeutic opportunities.
Oh S, Yeom J, Cho HJ, Kim JH, Yoon SJ, Kim H, Sa JK, Ju S, Lee H, Oh MJ, Lee W, Kwon Y, Li H, Choi S, Han JH, Chang JH, Choi E, Kim J, Her NG, Kim SH, Kang SG, Paek E, Nam DH, Lee C, Kim HS. Oh S, et al. Nat Commun. 2020 Jul 3;11(1):3288. doi: 10.1038/s41467-020-17139-y. Nat Commun. 2020. PMID: 32620753 Free PMC article. - A Reversible Shift of Driver Dependence from EGFR to Notch1 in Non-Small Cell Lung Cancer as a Cause of Resistance to Tyrosine Kinase Inhibitors.
Iommelli F, De Rosa V, Terlizzi C, Fonti R, Camerlingo R, Stoppelli MP, Stewart CA, Byers LA, Piwnica-Worms D, Del Vecchio S. Iommelli F, et al. Cancers (Basel). 2021 Apr 22;13(9):2022. doi: 10.3390/cancers13092022. Cancers (Basel). 2021. PMID: 33922104 Free PMC article. - Disrupting glioblastoma networks with tumor treating fields (TTFields) in in vitro models.
Schlieper-Scherf S, Hebach N, Hausmann D, Azorín DD, Hoffmann DC, Horschitz S, Maier E, Koch P, Karreman MA, Etminan N, Ratliff M. Schlieper-Scherf S, et al. J Neurooncol. 2024 Oct;170(1):139-151. doi: 10.1007/s11060-024-04786-0. Epub 2024 Aug 1. J Neurooncol. 2024. PMID: 39088157 Free PMC article.
References
- Hamburg MA & Collins FS The path to personalized medicine. N Engl J Med 363, 301–4 (2010). - PubMed
- Slamon DJ et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344, 783–92 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous