Multi-omic and multi-view clustering algorithms: review and cancer benchmark - PubMed (original) (raw)
Review
Multi-omic and multi-view clustering algorithms: review and cancer benchmark
Nimrod Rappoport et al. Nucleic Acids Res. 2018.
Erratum in
- Multi-omic and multi-view clustering algorithms: review and cancer benchmark.
Rappoport N, Shamir R. Rappoport N, et al. Nucleic Acids Res. 2019 Jan 25;47(2):1044. doi: 10.1093/nar/gky1226. Nucleic Acids Res. 2019. PMID: 30496480 Free PMC article. No abstract available.
Abstract
Recent high throughput experimental methods have been used to collect large biomedical omics datasets. Clustering of single omic datasets has proven invaluable for biological and medical research. The decreasing cost and development of additional high throughput methods now enable measurement of multi-omic data. Clustering multi-omic data has the potential to reveal further systems-level insights, but raises computational and biological challenges. Here, we review algorithms for multi-omics clustering, and discuss key issues in applying these algorithms. Our review covers methods developed specifically for omic data as well as generic multi-view methods developed in the machine learning community for joint clustering of multiple data types. In addition, using cancer data from TCGA, we perform an extensive benchmark spanning ten different cancer types, providing the first systematic comparison of leading multi-omics and multi-view clustering algorithms. The results highlight key issues regarding the use of single- versus multi-omics, the choice of clustering strategy, the power of generic multi-view methods and the use of approximated p-values for gauging solution quality. Due to the growing use of multi-omics data, we expect these issues to be important for future progress in the field.
Figures
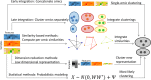
Figure 1.
Overview of multi-omics clustering approaches.
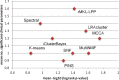
Figure 2.
Performance of the algorithms on ten multi-omics cancer datasets. For each plot, the x-axis measures the differential survival between clusters (–log10 of logrank’s test _P_-value), and the y-axis is the number of clinical parameters enriched in the clusters. Red vertical lines indicate the threshold for significantly different survival (_P_-value ≤ 0.05)
Figure 3.
Mean performance of the algorithms on ten multi-omics cancer datasets. The x-axis measures the differential survival between clusters (mean –log10 of logrank’s test _P_-value), and the y-axis is the mean number of clinical parameters enriched in the clusters.
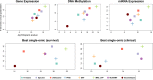
Figure 4.
Summarized performance of the algorithms across ten cancer datasets. For each plot, the x-axis measures the total differential prognosis between clusters (sum across all datasets of –log10 of logrank’s test _P_-value), and the y-axis is the total number of clinical parameters enriched in the clusters across all cancer types. (A–C) Results for single-omic datasets. (D) Results when each method uses the single omic that achieves the highest significance in survival. (E) Same with respect to enrichment of clinical labels.
Similar articles
- MONET: Multi-omic module discovery by omic selection.
Rappoport N, Safra R, Shamir R. Rappoport N, et al. PLoS Comput Biol. 2020 Sep 15;16(9):e1008182. doi: 10.1371/journal.pcbi.1008182. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32931516 Free PMC article. - Convex Multi-View Clustering Via Robust Low Rank Approximation With Application to Multi-Omic Data.
Shetta O, Niranjan M, Dasmahapatra S. Shetta O, et al. IEEE/ACM Trans Comput Biol Bioinform. 2022 Nov-Dec;19(6):3340-3352. doi: 10.1109/TCBB.2021.3122961. Epub 2022 Dec 8. IEEE/ACM Trans Comput Biol Bioinform. 2022. PMID: 34705655 - A multiobjective multi-view cluster ensemble technique: Application in patient subclassification.
Mitra S, Saha S. Mitra S, et al. PLoS One. 2019 May 23;14(5):e0216904. doi: 10.1371/journal.pone.0216904. eCollection 2019. PLoS One. 2019. PMID: 31120942 Free PMC article. - Evaluation of integrative clustering methods for the analysis of multi-omics data.
Chauvel C, Novoloaca A, Veyre P, Reynier F, Becker J. Chauvel C, et al. Brief Bioinform. 2020 Mar 23;21(2):541-552. doi: 10.1093/bib/bbz015. Brief Bioinform. 2020. PMID: 31220206 Review. - Multi-omic analysis tools for microbial metabolites prediction.
Wu S, Zhou H, Chen D, Lu Y, Li Y, Qiao J. Wu S, et al. Brief Bioinform. 2024 May 23;25(4):bbae264. doi: 10.1093/bib/bbae264. Brief Bioinform. 2024. PMID: 38859767 Free PMC article. Review.
Cited by
- Multi-view clustering for multi-omics data using unified embedding.
Mitra S, Saha S, Hasanuzzaman M. Mitra S, et al. Sci Rep. 2020 Aug 12;10(1):13654. doi: 10.1038/s41598-020-70229-1. Sci Rep. 2020. PMID: 32788601 Free PMC article. - The Need for Multi-Omics Biomarker Signatures in Precision Medicine.
Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. Olivier M, et al. Int J Mol Sci. 2019 Sep 26;20(19):4781. doi: 10.3390/ijms20194781. Int J Mol Sci. 2019. PMID: 31561483 Free PMC article. Review. - SADLN: Self-attention based deep learning network of integrating multi-omics data for cancer subtype recognition.
Sun Q, Cheng L, Meng A, Ge S, Chen J, Zhang L, Gong P. Sun Q, et al. Front Genet. 2023 Jan 4;13:1032768. doi: 10.3389/fgene.2022.1032768. eCollection 2022. Front Genet. 2023. PMID: 36685873 Free PMC article. - Multi-Omic Temporal Landscape of Plasma and Synovial Fluid-Derived Extracellular Vesicles Using an Experimental Model of Equine Osteoarthritis.
Anderson JR, Johnson E, Jenkins R, Jacobsen S, Green D, Walters M, Bundgaard L, Hausmans BAC, van den Akker G, Welting TJM, Chabronova A, Kharaz YA, Clarke EJ, James V, Peffers MJ. Anderson JR, et al. Int J Mol Sci. 2023 Oct 4;24(19):14888. doi: 10.3390/ijms241914888. Int J Mol Sci. 2023. PMID: 37834337 Free PMC article. - Non-Negative Symmetric Low-Rank Representation Graph Regularized Method for Cancer Clustering Based on Score Function.
Lu C, Wang J, Liu J, Zheng C, Kong X, Zhang X. Lu C, et al. Front Genet. 2020 Jan 22;10:1353. doi: 10.3389/fgene.2019.01353. eCollection 2019. Front Genet. 2020. PMID: 32038712 Free PMC article.
References
- Allison D.B., Cui X., Page G.P., Sabripour M.. Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 2006; 7:55–65. - PubMed
- Jain A.K., Murty M.N., Flynn P.J.. Data clustering: a review. ACM Comput. Surv. 1999; 31:264–323.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources