Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase - PubMed (original) (raw)
. 2018 Oct 18;8(1):15382.
doi: 10.1038/s41598-018-33731-1.
Amanda Borens 1, Álvaro Chiner-Oms 2, Paolo Miotto 3, Leonid Chindelevitch 4, Angela M Starks 5, Debra Hanna 1, Richard Liwski 1, Matteo Zignol 6, Christopher Gilpin 6, Stefan Niemann 7, Thomas Andreas Kohl 8, Robin M Warren 9, Derrick Crook 10, Sebastien Gagneux 11, Sven Hoffner 12, Camilla Rodrigues 13, Iñaki Comas 14, David M Engelthaler 15, David Alland 16, Leen Rigouts 17, Christoph Lange 18, Keertan Dheda 19, Rumina Hasan 20, Ruth McNerney 21, Daniela M Cirillo 3, Marco Schito 1, Timothy C Rodwell 22 23, James Posey 24
Affiliations
- PMID: 30337678
- PMCID: PMC6194142
- DOI: 10.1038/s41598-018-33731-1
Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase
Matthew Ezewudo et al. Sci Rep. 2018.
Erratum in
- Author Correction: Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase.
Ezewudo M, Borens A, Chiner-Oms Á, Miotto P, Chindelevitch L, Starks AM, Hanna D, Liwski R, Zignol M, Gilpin C, Niemann S, Kohl TA, Warren RM, Crook D, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, McNerney R, Cirillo DM, Schito M, Rodwell TC, Posey J. Ezewudo M, et al. Sci Rep. 2020 Feb 21;10(1):3531. doi: 10.1038/s41598-020-58955-y. Sci Rep. 2020. PMID: 32081980 Free PMC article.
Abstract
Drug-resistant tuberculosis poses a persistent public health threat. The ReSeqTB platform is a collaborative, curated knowledgebase, designed to standardize and aggregate global Mycobacterium tuberculosis complex (MTBC) variant data from whole genome sequencing (WGS) with phenotypic drug susceptibility testing (DST) and clinical data. We developed a unified analysis variant pipeline (UVP) ( https://github.com/CPTR-ReSeqTB/UVP ) to identify variants and assign lineage from MTBC sequence data. Stringent thresholds and quality control measures were incorporated in this open source tool. The pipeline was validated using a well-characterized dataset of 90 diverse MTBC isolates with conventional DST and DNA Sanger sequencing data. The UVP exhibited 98.9% agreement with the variants identified using Sanger sequencing and was 100% concordant with conventional methods of assigning lineage. We analyzed 4636 publicly available MTBC isolates in the ReSeqTB platform representing all seven major MTBC lineages. The variants detected have an above 94% accuracy of predicting drug based on the accompanying DST results in the platform. The aggregation of variants over time in the platform will establish confidence-graded mutations statistically associated with phenotypic drug resistance. These tools serve as critical reference standards for future molecular diagnostic assay developers, researchers, public health agencies and clinicians working towards the control of drug-resistant tuberculosis.
Conflict of interest statement
M. Schito, R. Liwski, A. Borens and M. Ezewudo reports grants from Bill & Melinda Gates Foundation, during the conduct of the study. Dr. D. Hanna reports grants from Bill & Melinda Gates Foundation, outside the submitted work. C. Lange reports personal fees from Chiesi, personal fees from Gilead, personal fees from Becton Dickinson, personal fees from Janssen, personal fees from Astra Zeneca, personal fees from Thermo Fisher Scientific, outside the submitted work. I. Comas reports personal fees from FIND Foundation for Innovative Diagnostics, within the scope of relevance to submitted work. D. Alland reports grants from Cepheid, other from Rutgers University Patent Pool, during the conduct of the study; In addition, D. Alland has a patent for primers and probes to detect drug resistance mutations issued. L. Rigouts reports other from FIND, during the conduct of the study. K. Dheda reports grants from FIND, grants and personal fees from ALERE, grants and personal fees from Oxford Immunotec, grants and personal fees from Cellestis (now Qiagen), grants from eNose Company, grants from Statens Serum Institut, grants and personal fees from bioMeriux, grants and personal fees from Cepheid, grants from Antrum Biotec, grants from Hain Lifescience, outside the submitted work; In addition, Dr. Dheda has a patent Characterization of novel TB-specific urinary biomarkers pending, a patent A smart mask for monitoring cough-related infectious diseases pending, and a patent device for diagnosing extrapulmonary tuberculosis (EPTB) issued. S. Niemann reports grants from German Center for Infection Research, during the conduct of the study and worked as consultant for FIND. T.C. Rodwell reports funding from NIH (NIAID) and FIND during the conduct of the study. Á. Chiner-Oms, P. Miotto, A.M. Starks, J. Posey, D. Crook, R.M. Warren, R. Hasan, M. Zignol, C. Gilpin, L. Chindelevitch, R. McNerney, D. Engelthaler, C. Rodrigues, S. Gagneux, D.M. Cirillo, S. Hoffner and T. A. Kohl have nothing to disclose.
Figures
Figure 1
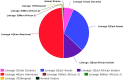
Distribution of isolates on ReSeqTB platform among the seven major lineages of the Mycobacterium tuberculosis complex (MTBC). This schematic shows the proportional representation of the major MTBC lineages in our dataset. The Euro American lineage has the most representation, but every other major lineage including Lineage 7 and animal strains are represented in the dataset.
Similar articles
- Diversified lineages and drug-resistance profiles of clinical isolates of Mycobacterium tuberculosis complex in Malaysia.
Noorizhab Fakhruzzaman MN, Abidin NZ, Aziz ZA, Lim WF, Richard JJ, Noorliza MN, Hani MH, Norhayati R, Zamzurina AB, Farida Zuraina MY, Hisyam MJ, Teh LK, Norazmi MN, Zaki MS. Noorizhab Fakhruzzaman MN, et al. Int J Mycobacteriol. 2019 Oct-Dec;8(4):320-328. doi: 10.4103/ijmy.ijmy_144_19. Int J Mycobacteriol. 2019. PMID: 31793500 - Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients.
Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, Rüsch-Gerdes S, Fattorini L, Oggioni MR, Cox H, Varaine F, Niemann S. Merker M, et al. PLoS One. 2013 Dec 6;8(12):e82551. doi: 10.1371/journal.pone.0082551. eCollection 2013. PLoS One. 2013. PMID: 24324807 Free PMC article. - Whole-Genome Sequencing for Drug Resistance Profile Prediction in Mycobacterium tuberculosis.
Gygli SM, Keller PM, Ballif M, Blöchliger N, Hömke R, Reinhard M, Loiseau C, Ritter C, Sander P, Borrell S, Collantes Loo J, Avihingsanon A, Gnokoro J, Yotebieng M, Egger M, Gagneux S, Böttger EC. Gygli SM, et al. Antimicrob Agents Chemother. 2019 Mar 27;63(4):e02175-18. doi: 10.1128/AAC.02175-18. Print 2019 Apr. Antimicrob Agents Chemother. 2019. PMID: 30718257 Free PMC article. - Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review.
Seifert M, Catanzaro D, Catanzaro A, Rodwell TC. Seifert M, et al. PLoS One. 2015 Mar 23;10(3):e0119628. doi: 10.1371/journal.pone.0119628. eCollection 2015. PLoS One. 2015. PMID: 25799046 Free PMC article. Review. - Use of whole genome sequencing in surveillance of drug resistant tuberculosis.
McNerney R, Zignol M, Clark TG. McNerney R, et al. Expert Rev Anti Infect Ther. 2018 May;16(5):433-442. doi: 10.1080/14787210.2018.1472577. Epub 2018 May 9. Expert Rev Anti Infect Ther. 2018. PMID: 29718745 Review.
Cited by
- Prediction of Acquired Antimicrobial Resistance for Multiple Bacterial Species Using Neural Networks.
Aytan-Aktug D, Clausen PTLC, Bortolaia V, Aarestrup FM, Lund O. Aytan-Aktug D, et al. mSystems. 2020 Jan 21;5(1):e00774-19. doi: 10.1128/mSystems.00774-19. mSystems. 2020. PMID: 31964771 Free PMC article. - Identification and Characterization of Mycobacterial Species Using Whole-Genome Sequences.
Riojas MA, Frank AM, Greenfield SR, King SP, Meehan CJ, Strong M, Wattam AR, Hazbón MH. Riojas MA, et al. Methods Mol Biol. 2021;2314:399-457. doi: 10.1007/978-1-0716-1460-0_19. Methods Mol Biol. 2021. PMID: 34235665 - The Bacteria Genome Pipeline (BAGEP): an automated, scalable workflow for bacteria genomes with Snakemake.
Olawoye IB, Frost SDW, Happi CT. Olawoye IB, et al. PeerJ. 2020 Oct 27;8:e10121. doi: 10.7717/peerj.10121. eCollection 2020. PeerJ. 2020. PMID: 33194387 Free PMC article. - CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database.
Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, Baker SJC, Dave M, McCarthy MC, Mukiri KM, Nasir JA, Golbon B, Imtiaz H, Jiang X, Kaur K, Kwong M, Liang ZC, Niu KC, Shan P, Yang JYJ, Gray KL, Hoad GR, Jia B, Bhando T, Carfrae LA, Farha MA, French S, Gordzevich R, Rachwalski K, Tu MM, Bordeleau E, Dooley D, Griffiths E, Zubyk HL, Brown ED, Maguire F, Beiko RG, Hsiao WWL, Brinkman FSL, Van Domselaar G, McArthur AG. Alcock BP, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D690-D699. doi: 10.1093/nar/gkac920. Nucleic Acids Res. 2023. PMID: 36263822 Free PMC article. - A Bioinformatics Whole-Genome Sequencing Workflow for Clinical Mycobacterium tuberculosis Complex Isolate Analysis, Validated Using a Reference Collection Extensively Characterized with Conventional Methods and In Silico Approaches.
Bogaerts B, Delcourt T, Soetaert K, Boarbi S, Ceyssens PJ, Winand R, Van Braekel J, De Keersmaecker SCJ, Roosens NHC, Marchal K, Mathys V, Vanneste K. Bogaerts B, et al. J Clin Microbiol. 2021 May 19;59(6):e00202-21. doi: 10.1128/JCM.00202-21. Print 2021 May 19. J Clin Microbiol. 2021. PMID: 33789960 Free PMC article.
References
- WHO. Global tuberculosis control (2011).
- WHO. Global tuberculosis report (2015).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases