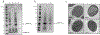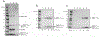In Vitro Assembly of Diverse Bacterial Microcompartment Shell Architectures - PubMed (original) (raw)
In Vitro Assembly of Diverse Bacterial Microcompartment Shell Architectures
Andrew R Hagen et al. Nano Lett. 2018.
Abstract
Bacterial microcompartments (BMCs) are organelles composed of a selectively permeable protein shell that encapsulates enzymes involved in CO2 fixation (carboxysomes) or carbon catabolism (metabolosomes). Confinement of sequential reactions by the BMC shell presumably increases the efficiency of the pathway by reducing the crosstalk of metabolites, release of toxic intermediates, and accumulation of inhibitory products. Because BMCs are composed entirely of protein and self-assemble, they are an emerging platform for engineering nanoreactors and molecular scaffolds. However, testing designs for assembly and function through in vivo expression is labor-intensive and has limited the potential of BMCs in bioengineering. Here, we developed a new method for in vitro assembly of defined nanoscale BMC architectures: shells and nanotubes. By inserting a "protecting group", a short ubiquitin-like modifier (SUMO) domain, self-assembly of shell proteins in vivo was thwarted, enabling preparation of concentrates of shell building blocks. Addition of the cognate protease removes the SUMO domain and subsequent mixing of the constituent shell proteins in vitro results in the self-assembly of three types of supramolecular architectures: a metabolosome shell, a carboxysome shell, and a BMC protein-based nanotube. We next applied our method to generate a metabolosome shell engineered with a hyper-basic luminal surface, allowing for the encapsulation of biotic or abiotic cargos functionalized with an acidic accessory group. This is the first demonstration of using charge complementarity to encapsulate diverse cargos in BMC shells. Collectively, our work provides a generally applicable method for in vitro assembly of natural and engineered BMC-based architectures.
Keywords: Bacterial microcompartments; electrostatic-based encapsulation; in vitro supramolecular self-assembly; nanotubes; protein engineering; shells; structural biology.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
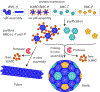
Figure 1.
Schematic of the macromolecular protecting group strategy for in vitro assembly of BMC architectures.
Figure 2.
Characterization of SUMOylated BMC-H proteins. (a) SDS-PAGE analysis of fractionated whole cell lysates of Escherichia coli strains expressing wildtype BMC-HHO (lanes 1, 2) and SUMOBMC-HHO (lanes 3, 4). (b) SDS-PAGE analysis of SUMOBMC-HRmm. For a. and b., soluble fractions are indicated with an “S” and insoluble fractions with “I,” and asterisks denote soluble protein of interest. (c) Transmission electron micrographs of thin sections of E. coli strains expressing wildtype BMC-H (upper left panel), SUMOBMC-HHO (lower left panel), WT BMC-HRmm (upper right panel), and SUMOBMC-HRmm (lower right panel). Depicted results are representative of multiple fields/cells. Scale bars represent 200 nm.
Figure 3.
Transmission electron microscopy of various in vitro assembled nanoarchitectures. (a) In vitro assembled BMC-HRmm nanotubes. (b) In vitro assembled minimal HO shells. (c) In vitro assembled Halo shells (see also Figure S6). Scale bars = 100 nm; depicted results are representative of multiple fields.
Figure 4.
SDS-PAGE analysis of in vitro assembly of minimal HO shells. (a) Shell assembly using CAP (dashed line indicates cropped out lanes of irrelevant samples). Lane 1, 2 and 3 contains the neat assembly reaction, StrepTrap flow-through, and StrepTrap eluate, respectively. (b) BMC HO trimer sensitivity analysis showing five-fold concentrated eluates. Stoichiometry of BMC-T1HO trimer used is shown beneath each lane. (c) Shell assembly time course (five-fold concentrated eluates; 3x BMC-T1HO trimer stoichiometry used). Time between addition of MBP-Ulp protease and application of reaction to StrepTrap column is indicated below each lane. See Figure S9 for uncropped images of Figures 4b and 4c.
Figure 5.
Characterization of the electrostatic-based encapsulation of cargo. (a) Cutaway of the full HO shell structure (PDB ID: 5V74) incorporating the modelled electrostatic potential map of three BMC-HHO (left) and BMC-H+HO (right) hexamers. The electrostatic potential surface is colored from −5kT/e (red) to +5kT/e (blue). (b) Representative TEM image of purified mHO+ + RraB-GFP (scale bar = 100 nm). Inset is an enlarged view of the shells. See Figure S10c for a representative TEM micrograph of mHO+ + GFP (c) SDS-PAGE analysis of purified in vitro mHO/mHO+ shell assemblies with RraB-GFP or GFP. A comparable amount of protein was loaded in all the lanes, 1.2 – 1.9 μg; see Figure S11 for the DLS analysis of these assemblies. (d) Fluorescence emission spectra of purified of mHO(+) shell assemblies with RraB-GFP or GFP. The error bars in the emission spectra plots are standard deviations from the average fluorescence emission at each wavelength measured for three independent purified reactions. (e) Various TEM images of mHO+ copurified with 5 nm Au-COOH particles (scale bars = 50 nm).
Similar articles
- Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells.
Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. Cai F, et al. ACS Synth Biol. 2015 Apr 17;4(4):444-53. doi: 10.1021/sb500226j. Epub 2014 Aug 27. ACS Synth Biol. 2015. PMID: 25117559 - The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment.
Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. Cai F, et al. J Biol Chem. 2013 May 31;288(22):16055-63. doi: 10.1074/jbc.M113.456897. Epub 2013 Apr 9. J Biol Chem. 2013. PMID: 23572529 Free PMC article. - Engineering nanoreactors using bacterial microcompartment architectures.
Plegaria JS, Kerfeld CA. Plegaria JS, et al. Curr Opin Biotechnol. 2018 Jun;51:1-7. doi: 10.1016/j.copbio.2017.09.005. Epub 2017 Oct 13. Curr Opin Biotechnol. 2018. PMID: 29035760 Free PMC article. Review. - The Plasticity of Molecular Interactions Governs Bacterial Microcompartment Shell Assembly.
Greber BJ, Sutter M, Kerfeld CA. Greber BJ, et al. Structure. 2019 May 7;27(5):749-763.e4. doi: 10.1016/j.str.2019.01.017. Epub 2019 Mar 1. Structure. 2019. PMID: 30833088 Free PMC article. - Dynamic structural determinants in bacterial microcompartment shells.
Trettel DS, Kerfeld CA, Gonzalez-Esquer CR. Trettel DS, et al. Curr Opin Microbiol. 2024 Aug;80:102497. doi: 10.1016/j.mib.2024.102497. Epub 2024 Jun 21. Curr Opin Microbiol. 2024. PMID: 38909546 Review.
Cited by
- Characterization of a widespread sugar phosphate-processing bacterial microcompartment.
Dwyer ME, Sutter M, Kerfeld CA. Dwyer ME, et al. Commun Biol. 2024 Nov 24;7(1):1562. doi: 10.1038/s42003-024-07287-y. Commun Biol. 2024. PMID: 39580597 Free PMC article. - Rubisco packaging and stoichiometric composition of a native β-carboxysome.
Sun Y, Sheng Y, Ni T, Ge X, Sarsby J, Brownridge PJ, Li K, Hardenbrook N, Dykes GF, Rockliffe N, Eyers CE, Zhang P, Liu LN. Sun Y, et al. bioRxiv [Preprint]. 2024 Sep 21:2024.09.20.614183. doi: 10.1101/2024.09.20.614183. bioRxiv. 2024. PMID: 39345498 Free PMC article. Updated. Preprint. - Comparative Pore Structure and Dynamics for Bacterial Microcompartment Shell Protein Assemblies in Sheets or Shells.
Raza S, Sarkar D, Chan LJG, Mae J, Sutter M, Petzold CJ, Kerfeld CA, Ralston CY, Gupta S, Vermaas JV. Raza S, et al. ACS Omega. 2024 Jul 18;9(33):35503-35514. doi: 10.1021/acsomega.4c02406. eCollection 2024 Aug 20. ACS Omega. 2024. PMID: 39184480 Free PMC article. - Bacterial microcompartments as a next-generation metabolic engineering tool: utilizing nature's solution for confining challenging catabolic pathways.
Doron L, Kerfeld CA. Doron L, et al. Biochem Soc Trans. 2024 Jun 26;52(3):997-1010. doi: 10.1042/BST20230229. Biochem Soc Trans. 2024. PMID: 38813858 Free PMC article. Review. - Stimulus-responsive assembly of nonviral nucleocapsids.
Hori M, Steinauer A, Tetter S, Hälg J, Manz EM, Hilvert D. Hori M, et al. Nat Commun. 2024 Apr 27;15(1):3576. doi: 10.1038/s41467-024-47808-1. Nat Commun. 2024. PMID: 38678040 Free PMC article.
References
- Sutter M; Boehringer D; Gutmann S; Gunther S; Prangishvili D; Loessner MJ; Stetter KO; Weber-Ban E; Ban N Nat Struct Mol Biol 2008, 15, (9), 939–47. - PubMed
- Giessen TW; Silver PA Nat Microbiol 2017, 2, 17029. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources