Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis - PubMed (original) (raw)
Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis
Robert N Kirchdoerfer et al. Sci Rep. 2018.
Erratum in
- Publisher Correction: Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB. Kirchdoerfer RN, et al. Sci Rep. 2018 Dec 10;8(1):17823. doi: 10.1038/s41598-018-36918-8. Sci Rep. 2018. PMID: 30531867 Free PMC article.
Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 as a highly transmissible pathogenic human betacoronavirus. The viral spike glycoprotein (S) utilizes angiotensin-converting enzyme 2 (ACE2) as a host protein receptor and mediates fusion of the viral and host membranes, making S essential to viral entry into host cells and host species tropism. As SARS-CoV enters host cells, the viral S is believed to undergo a number of conformational transitions as it is cleaved by host proteases and binds to host receptors. We recently developed stabilizing mutations for coronavirus spikes that prevent the transition from the pre-fusion to post-fusion states. Here, we present cryo-EM analyses of a stabilized trimeric SARS-CoV S, as well as the trypsin-cleaved, stabilized S, and its interactions with ACE2. Neither binding to ACE2 nor cleavage by trypsin at the S1/S2 cleavage site impart large conformational changes within stabilized SARS-CoV S or expose the secondary cleavage site, S2'.
Conflict of interest statement
R.N.K., N.W., J.P., H.L.T., C.A.C., K.S.C., B.S.G., J.S.M. and A.B.W. are inventors on US patent Application No. 62/412,703, entitled “Prefusion Coronavirus Spike Proteins and Their Use”. D.W. has no competing interests.
Figures
Figure 1
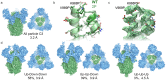
Structure of the SARS-CoV S 2P ectotodomain. (a) The C3 symmetrized reconstruction of all particles within the dataset resembles that of other betacoronaviruses. Side and membrane-distal top views (90° rotation about x-axis) are shown. (b) Coordinate models derived from cryo-EM reconstructions of the wild-type SARS-CoV S ectodomain (5X58.pdb, dark green) and the prefusion stabilized SARS-CoV S 2P ectodomain (6CRV.pdb light green) adopt identical conformations near the 2P mutation site. Coordinate models derived from C3 symmetry cryo-EM reconstructions are shown. (c) There is highly featured density in the region containing the 2P mutation site. (d) Classification of heterogeneity within the S1 RBD reveals a distribution of RBD configurations with the single-‘up’ conformation being most prevalent. S1 regions are shown in blue and S2 regions are shown in green.
Figure 2
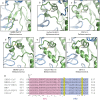
Comparison of putative bi-partite fusion peptide regions. (a) The SARS-CoV S FP2 region of the bi-partite fusion peptide adopts a conformation distinct from equivalent regions in (b) alpha- (HuCoV-NL63, 5SZS.pdb) and (c) deltacoronavirus spikes (PDCV S, 6BFU.pdb) as well as other (d–f) betacoronaviruses (MERS-CoV S, 5W9I.pdb, HuCoV-HKU1 S, 5I08.pdb, MHV S, 3JCL.pdb). Subunits are colored as in Fig. 1. (g) Sequence alignment of the fusion peptide regions of coronavirus spikes highlighting the FP1 and FP2 regions as well as the cysteine residues involved in disulfide bond formation indicated in panels a–f. Sequence alignment was prepared with Clustal Omega.
Figure 3
Compositional and conformational heterogeneity of ACE2-bound SARS-CoV spikes. Classification of heterogeneity within the SARS-CoV S – ACE2 complex reveals that 55% of the particles are not bound by ACE2. Of those particles which are bound by ACE2, each configuration of the S1 RBD is observed with classes containing triple ‘up’ conformation S1 RBD being the most poorly represented. The labeled description of each class begins at the lower RBD when viewed at the membrane distal apex and proceeds clockwise. Side views and membrane-distal top views are shown for each reconstruction. S1 regions are shown in blue, S2 regions are shown in green and soluble ACE2 is shown in orange.
Figure 4
ACE2-receptor binding and induced conformational changes. (a) ACE2 binds the SARS-CoV S1 RBD in an ‘up’ conformation. (b) Focused refinement was used on the S1 RBD – ACE2 portion of the particles to improve the density of this flexible region. (c) Rigid-body fitting of the SARS-CoV S1 RBD – ACE2 complex crystal structure (2AJF.pdb) indicates that the crystal structure is representative of the ACE2-bound trimeric spike. Comparison of the d) SARS-CoV S 2P central helices with the (e) SARS-CoV S 2P central helix which has been uncapped by an ACE2-bound S1 RBD demonstrates a transition to a short 310-helix. (f) The ACE2-bound SARS-CoV S 2P central helix demonstrates good density in this region. Colors are as in Fig. 3.
Figure 5
Trypsin cleavage does not impart conformational changes in prefusion SARS-CoV S 2P spike. (a) An all particle reconstruction of trypsin-cleaved SARS-CoV S 2P with C3 symmetry. (b) Comparison of the trypsin-cleaved and uncleaved SARS-CoV S 2P coordinate models reveals no differences in protein structure near the cleavage site. The last residue in S1 (T662) and the first residue in S2 (K672) visualized in the coordinates are labeled. (c) Classification of the heterogeneous S1 RBD in the trypsin-cleaved S shows a predominance of the single-‘up’ conformation as well as the observation of an all-‘down’ conformation not seen in the uncleaved S 2P sample. Colors are as in Fig. 1.
Figure 6
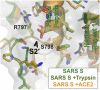
S2′ secondary cleavage site is occluded. Superimposition of the SARS-CoV S 2P, uncleaved, cleaved and ACE2-bound structures reveals no structural changes within this region. The S2′ site after amino acid R797 is shielded from cleavage by an adjacent loop and the fusion peptide which packs against an N-terminal region of S2 HR1.
Similar articles
- Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2.
Song W, Gui M, Wang X, Xiang Y. Song W, et al. PLoS Pathog. 2018 Aug 13;14(8):e1007236. doi: 10.1371/journal.ppat.1007236. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30102747 Free PMC article. - SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. Wrobel AG, et al. Nat Struct Mol Biol. 2020 Aug;27(8):763-767. doi: 10.1038/s41594-020-0468-7. Epub 2020 Jul 9. Nat Struct Mol Biol. 2020. PMID: 32647346 Free PMC article. - TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein.
Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. Heurich A, et al. J Virol. 2014 Jan;88(2):1293-307. doi: 10.1128/JVI.02202-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227843 Free PMC article. - Coronavirus Spike Protein and Tropism Changes.
Hulswit RJ, de Haan CA, Bosch BJ. Hulswit RJ, et al. Adv Virus Res. 2016;96:29-57. doi: 10.1016/bs.aivir.2016.08.004. Epub 2016 Sep 13. Adv Virus Res. 2016. PMID: 27712627 Free PMC article. Review. - COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2.
Mittal A, Manjunath K, Ranjan RK, Kaushik S, Kumar S, Verma V. Mittal A, et al. PLoS Pathog. 2020 Aug 21;16(8):e1008762. doi: 10.1371/journal.ppat.1008762. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32822426 Free PMC article. Review.
Cited by
- Thermodynamic equilibrium dose-response models for MERS-CoV infection reveal a potential protective role of human lung mucus but not for SARS-CoV-2.
Gale P. Gale P. Microb Risk Anal. 2020 Dec;16:100140. doi: 10.1016/j.mran.2020.100140. Epub 2020 Sep 19. Microb Risk Anal. 2020. PMID: 32984489 Free PMC article. - Biophysical and Biochemical Characterization of the Receptor Binding Domain of SARS-CoV-2 Variants.
Khatri R, Parray HA, Siddiqui G, Chiranjivi AK, Raj S, Kaul R, Maithil V, Samal S, Ahmed S. Khatri R, et al. Protein J. 2022 Oct;41(4-5):457-467. doi: 10.1007/s10930-022-10073-6. Epub 2022 Sep 1. Protein J. 2022. PMID: 36048314 Free PMC article. - Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates.
He L, Lin X, Wang Y, Abraham C, Sou C, Ngo T, Zhang Y, Wilson IA, Zhu J. He L, et al. Sci Adv. 2021 Mar 19;7(12):eabf1591. doi: 10.1126/sciadv.abf1591. Print 2021 Mar. Sci Adv. 2021. PMID: 33741598 Free PMC article. - In silico anti-viral assessment of phytoconstituents in a traditional (Siddha Medicine) polyherbal formulation - Targeting Mpro and pan-coronavirus post-fusion Spike protein.
Mandal SK, Rehman MM, Katyal A, Rajvanshi K, Kannan M, Garg M, Murugesan S, Deepa PR. Mandal SK, et al. J Tradit Complement Med. 2023 Jul 13;14(1):55-69. doi: 10.1016/j.jtcme.2023.07.004. eCollection 2024 Jan. J Tradit Complement Med. 2023. PMID: 38223813 Free PMC article. - Mutations in membrane-fusion subunit of spike glycoprotein play crucial role in the recent outbreak of COVID-19.
Podder S, Ghosh A, Ghosh T. Podder S, et al. J Med Virol. 2021 May;93(5):2790-2798. doi: 10.1002/jmv.26598. Epub 2021 Feb 23. J Med Virol. 2021. PMID: 33090493 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous