GENCODE reference annotation for the human and mouse genomes - PubMed (original) (raw)
. 2019 Jan 8;47(D1):D766-D773.
doi: 10.1093/nar/gky955.
Mark Diekhans 2, Anne-Maud Ferreira 3, Rory Johnson 4 5, Irwin Jungreis 6 7, Jane Loveland 1, Jonathan M Mudge 1, Cristina Sisu 8 9, James Wright 10, Joel Armstrong 2, If Barnes 1, Andrew Berry 1, Alexandra Bignell 1, Silvia Carbonell Sala 11, Jacqueline Chrast 3, Fiona Cunningham 1, Tomás Di Domenico 12, Sarah Donaldson 1, Ian T Fiddes 2, Carlos García Girón 1, Jose Manuel Gonzalez 1, Tiago Grego 1, Matthew Hardy 1, Thibaut Hourlier 1, Toby Hunt 1, Osagie G Izuogu 1, Julien Lagarde 11, Fergal J Martin 1, Laura Martínez 12, Shamika Mohanan 1, Paul Muir 13 14, Fabio C P Navarro 8, Anne Parker 1, Baikang Pei 8, Fernando Pozo 12, Magali Ruffier 1, Bianca M Schmitt 1, Eloise Stapleton 1, Marie-Marthe Suner 1, Irina Sycheva 1, Barbara Uszczynska-Ratajczak 15, Jinuri Xu 8, Andrew Yates 1, Daniel Zerbino 1, Yan Zhang 8 16, Bronwen Aken 1, Jyoti S Choudhary 10, Mark Gerstein 8 17 18, Roderic Guigó 11 19, Tim J P Hubbard 20, Manolis Kellis 6 7, Benedict Paten 2, Alexandre Reymond 3, Michael L Tress 12, Paul Flicek 1
Affiliations
- PMID: 30357393
- PMCID: PMC6323946
- DOI: 10.1093/nar/gky955
GENCODE reference annotation for the human and mouse genomes
Adam Frankish et al. Nucleic Acids Res. 2019.
Abstract
The accurate identification and description of the genes in the human and mouse genomes is a fundamental requirement for high quality analysis of data informing both genome biology and clinical genomics. Over the last 15 years, the GENCODE consortium has been producing reference quality gene annotations to provide this foundational resource. The GENCODE consortium includes both experimental and computational biology groups who work together to improve and extend the GENCODE gene annotation. Specifically, we generate primary data, create bioinformatics tools and provide analysis to support the work of expert manual gene annotators and automated gene annotation pipelines. In addition, manual and computational annotation workflows use any and all publicly available data and analysis, along with the research literature to identify and characterise gene loci to the highest standard. GENCODE gene annotations are accessible via the Ensembl and UCSC Genome Browsers, the Ensembl FTP site, Ensembl Biomart, Ensembl Perl and REST APIs as well as https://www.gencodegenes.org.
Figures
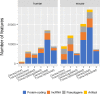
Figure 1.
New and updated manually annotated genes and transcripts from July 2016 to June 2018. For both human (left) and mouse (right) the numbers of completely new genes and transcripts, updated genes and transcripts and the total number of manually added or edited genes and transcripts for each of four broad categories of annotation. A new gene annotation can represent a completely de novo locus with no overlap with pre-existing annotation or the reclassification of an existing complex locus into multiple loci to better represent the biology of the locus inferred from transcriptomic and/or proteomic data. A new transcript represents the annotation of a unique exon-intron structure, including novel alternative splicing at an annotated locus. Updated genes and transcripts represent pre-existing loci or transcript models that have been edited to improve the representation of biotype (e.g. changed from lncRNA to protein-coding) or structure (e.g. by extension, addition of novel exons).
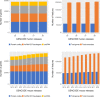
Figure 2.
Annotation statistics for human and mouse GENCODE releases from July 2016 to June 2018, encompassing human releases GENCODE 25–28 and mouse releases M10 to M18. The panels on the left show the total number of genes by broad biotype (protein-coding, lncRNA, pseudogene and sncRNA) for each release for human and mouse respectively and panels on the right show the total numbers of genes and transcripts of all biotypes.
Similar articles
- GENCODE 2021.
Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland JE, Mudge JM, Sisu C, Wright JC, Armstrong J, Barnes I, Berry A, Bignell A, Boix C, Carbonell Sala S, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Howe KL, Hunt T, Izuogu OG, Johnson R, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Riera FC, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Wolf MY, Xu J, Yang YT, Yates A, Zerbino D, Zhang Y, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Tress ML, Flicek P. Frankish A, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D916-D923. doi: 10.1093/nar/gkaa1087. Nucleic Acids Res. 2021. PMID: 33270111 Free PMC article. - GENCODE: reference annotation for the human and mouse genomes in 2023.
Frankish A, Carbonell-Sala S, Diekhans M, Jungreis I, Loveland JE, Mudge JM, Sisu C, Wright JC, Arnan C, Barnes I, Banerjee A, Bennett R, Berry A, Bignell A, Boix C, Calvet F, Cerdán-Vélez D, Cunningham F, Davidson C, Donaldson S, Dursun C, Fatima R, Giorgetti S, Giron CG, Gonzalez JM, Hardy M, Harrison PW, Hourlier T, Hollis Z, Hunt T, James B, Jiang Y, Johnson R, Kay M, Lagarde J, Martin FJ, Gómez LM, Nair S, Ni P, Pozo F, Ramalingam V, Ruffier M, Schmitt BM, Schreiber JM, Steed E, Suner MM, Sumathipala D, Sycheva I, Uszczynska-Ratajczak B, Wass E, Yang YT, Yates A, Zafrulla Z, Choudhary JS, Gerstein M, Guigo R, Hubbard TJP, Kellis M, Kundaje A, Paten B, Tress ML, Flicek P. Frankish A, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D942-D949. doi: 10.1093/nar/gkac1071. Nucleic Acids Res. 2023. PMID: 36420896 Free PMC article. - GENCODE: the reference human genome annotation for The ENCODE Project.
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. Harrow J, et al. Genome Res. 2012 Sep;22(9):1760-74. doi: 10.1101/gr.135350.111. Genome Res. 2012. PMID: 22955987 Free PMC article. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review. - Assembly, Annotation, and Comparative Genomics in PATRIC, the All Bacterial Bioinformatics Resource Center.
Wattam AR, Brettin T, Davis JJ, Gerdes S, Kenyon R, Machi D, Mao C, Olson R, Overbeek R, Pusch GD, Shukla MP, Stevens R, Vonstein V, Warren A, Xia F, Yoo H. Wattam AR, et al. Methods Mol Biol. 2018;1704:79-101. doi: 10.1007/978-1-4939-7463-4_4. Methods Mol Biol. 2018. PMID: 29277864 Review.
Cited by
- A multiplex single-cell RNA-Seq pharmacotranscriptomics pipeline for drug discovery.
Dini A, Barker H, Piki E, Sharma S, Raivola J, Murumägi A, Ungureanu D. Dini A, et al. Nat Chem Biol. 2024 Oct 31. doi: 10.1038/s41589-024-01761-8. Online ahead of print. Nat Chem Biol. 2024. PMID: 39482470 - Transcript Identification Through Long-Read Sequencing.
Seki M, Oka M, Xu L, Suzuki A, Suzuki Y. Seki M, et al. Methods Mol Biol. 2021;2284:531-541. doi: 10.1007/978-1-0716-1307-8_29. Methods Mol Biol. 2021. PMID: 33835462 - Genome-wide DNA Methylation Signatures Are Determined by DNMT3A/B Sequence Preferences.
Mao SQ, Cuesta SM, Tannahill D, Balasubramanian S. Mao SQ, et al. Biochemistry. 2020 Jul 14;59(27):2541-2550. doi: 10.1021/acs.biochem.0c00339. Epub 2020 Jun 28. Biochemistry. 2020. PMID: 32543182 Free PMC article. - MTGR1 is required to maintain small intestinal stem cell populations.
Short SP, Brown RE, Chen Z, Pilat JM, McElligott BA, Meenderink LM, Bickart AC, Blunt KM, Jacobse J, Wang J, Simmons AJ, Xu Y, Yang Y, Parang B, Choksi YA, Goettel JA, Lau KS, Hiebert SW, Williams CS. Short SP, et al. Cell Death Differ. 2024 Sep;31(9):1170-1183. doi: 10.1038/s41418-024-01346-x. Epub 2024 Jul 25. Cell Death Differ. 2024. PMID: 39048708 Free PMC article. - A mouse tissue atlas of small noncoding RNA.
Isakova A, Fehlmann T, Keller A, Quake SR. Isakova A, et al. Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25634-25645. doi: 10.1073/pnas.2002277117. Epub 2020 Sep 25. Proc Natl Acad Sci U S A. 2020. PMID: 32978296 Free PMC article.
References
- ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004; 306:636–640. - PubMed
Publication types
MeSH terms
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- U41 HG007234/HG/NHGRI NIH HHS/United States
- WT200990/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- WT108749/Z/15/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases