Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae - PubMed (original) (raw)
Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae
Margaret M C Lam et al. Genome Med. 2018.
Abstract
Background: Klebsiella pneumoniae is a recognised agent of multidrug-resistant (MDR) healthcare-associated infections; however, individual strains vary in their virulence potential due to the presence of mobile accessory genes. In particular, gene clusters encoding the biosynthesis of siderophores aerobactin (iuc) and salmochelin (iro) are associated with invasive disease and are common amongst hypervirulent K. pneumoniae clones that cause severe community-associated infections such as liver abscess and pneumonia. Concerningly, iuc has also been reported in MDR strains in the hospital setting, where it was associated with increased mortality, highlighting the need to understand, detect and track the mobility of these virulence loci in the K. pneumoniae population.
Methods: Here, we examined the genetic diversity, distribution and mobilisation of iuc and iro loci amongst 2503 K. pneumoniae genomes using comparative genomics approaches and developed tools for tracking them via genomic surveillance.
Results: Iro and iuc were detected at low prevalence (< 10%). Considerable genetic diversity was observed, resolving into five iro and six iuc lineages that show distinct patterns of mobilisation and dissemination in the K. pneumoniae population. The major burden of iuc and iro amongst the genomes analysed was due to two linked lineages (iuc1/iro1 74% and iuc2/iro2 14%), each carried by a distinct non-self-transmissible IncFIBK virulence plasmid type that we designate KpVP-1 and KpVP-2. These dominant types also carry hypermucoidy (rmpA) determinants and include all previously described virulence plasmids of K. pneumoniae. The other iuc and iro lineages were associated with diverse plasmids, including some carrying IncFII conjugative transfer regions and some imported from Escherichia coli; the exceptions were iro3 (mobilised by ICEKp1) and iuc4 (fixed in the chromosome of K. pneumoniae subspecies rhinoscleromatis). Iro/iuc mobile genetic elements (MGEs) appear to be stably maintained at high frequency within known hypervirulent strains (ST23, ST86, etc.) but were also detected at low prevalence in others such as MDR strain ST258.
Conclusions: Iuc and iro are mobilised in K. pneumoniae via a limited number of MGEs. This study provides a framework for identifying and tracking these important virulence loci, which will be important for genomic surveillance efforts including monitoring for the emergence of hypervirulent MDR K. pneumoniae strains.
Keywords: Aerobactin; Genomic surveillance; Hypervirulence; Invasive disease; Klebsiella pneumoniae; Plasmids; Salmochelin; Virulence; Virulence plasmids.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
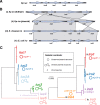
Aerobactin and salmochelin locus variants found in Klebsiella pnuemoniae. a A single aerobactin (iuc) locus structure was found in K. pneumoniae. b Four different structures of the salmochelin (iro) locus were found in K. pneumoniae (i–iv). Note two of these are typical of structures found in other species (iii in Enterobacter cloacae, iv in Escherichia coli). c Maximum likelihood phylogenetic trees inferred from iuc and iro sequence types (AbSTs and SmSTs) identified in K. pneumoniae genomes. Phylogenetic lineages discussed in the text are labelled and their mobility indicated; nucleotide divergence within and between lineages is given in Additional files 8 and 9. Iro locus structures associated with each lineage are labelled i–iv, as defined in panel b
Fig. 2
Plasmid variants associated with different iro and/or iuc lineages identified amongst K. pneumoniae. a Clustering of the 12 reference plasmids based on gene content, annotated with the presence of iuc and iro lineages (coloured as in panel b and Fig. 1c), rmpA, IncFIBK, IncFIB, IncFII and/or other plasmid replicon types. b Gene content matrix for reference plasmids; columns correspond to protein-coding sequences that are > 10% divergent from one another. IncFII tra-trb conjugal transfer region genes are coloured blue, to highlight the divergent forms of this region and labelled with the closest IncFII type as detected by PlasmidFinder. c Genetic maps for the reference plasmids. The positions of key loci involved in core plasmid functions (bold), virulence (iro highlighted in yellow, iuc in dark orange and other loci involved in iron acquisition/transport in light orange) and antimicrobial resistance are indicated. Grey shading indicates homology blocks sharing > 60% nucleotide identity
Fig. 3
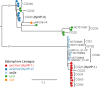
Maximum likelihood phylogeny of representative IncFIBK replicon sequences from isolates with iuc/iro plasmids. Each tip represents a unique IncFIBK replicon sequence (spanning repA, oriT, sopAB), coloured according to the iro/iuc lineage carried by the corresponding isolates as per inset legend. IncFIBK sequences found in the representative plasmid sequences (shown in Fig. 2 and listed in Additional file 2) are labelled; tips/subclades are also annotated to indicate those found in common clonal groups (CG; see Fig. 5)
Fig. 4
Conservation of reference plasmid genes amongst isolates with plasmid-associated iuc/iro lineages. Cells show circularised heatmaps indicating the frequency of each gene in a given reference plasmid (column), amongst isolates that contain a given iro and/or iuc lineage (row). Around each circle, genes are ordered by their order in the corresponding reference plasmid. Percentages in the middle of each cell indicate the mean coverage of the reference plasmid sequence (column), amongst isolates belonging to each iro/iuc lineage (row); bold labels and boxes highlight groups of isolates carrying the same iuc/iro lineage as the reference plasmid. ‘*’ indicates the two plasmids represented by incomplete plasmid sequences
Fig. 5
Distribution of plasmid and chromosomal variants of iro and iuc and capsule locus (KL) types amongst K. pneumoniae clones. Rows indicate sequence types (STs, as labelled) that contain ≥ 1 genome in which iro and/or iuc was detected; vertical lines indicate STs belonging to the same clonal group (CG) as labelled. Pie charts indicate prevalence of iro and/or iuc within common K. pneumoniae lineages. The detection of individual iro and iuc lineages within each K. pneumoniae ST is indicated in the grid, coloured as per Fig. 1. Bar plots indicate sample size (number of genomes per ST; note log10 scale). Heatmap on the right indicates prevalence of capsule (K) locus types in each K. pneumoniae ST, coloured as per inset legend. Individual columns are included for K types that are common amongst virulent clones; where other K types were detected, these are represented in the ‘other’ column, and the relevant K type for that ST is labelled to the right
Similar articles
- Genetic diversity and evolution of the virulence plasmids encoding aerobactin and salmochelin in Klebsiella pneumoniae.
Tian D, Wang M, Zhou Y, Hu D, Ou HY, Jiang X. Tian D, et al. Virulence. 2021 Dec;12(1):1323-1333. doi: 10.1080/21505594.2021.1924019. Virulence. 2021. PMID: 33970792 Free PMC article. - Co-Occurrence of Rare ArmA-, RmtB-, and KPC-2-Encoding Multidrug-Resistant Plasmids and Hypervirulence iuc Operon in ST11-KL47 Klebsiella pneumoniae.
Zhou Y, Ai W, Guo Y, Wu X, Wang B, Xu Y, Rao L, Zhao H, Wang X, Yu F. Zhou Y, et al. Microbiol Spectr. 2022 Apr 27;10(2):e0237121. doi: 10.1128/spectrum.02371-21. Epub 2022 Mar 24. Microbiol Spectr. 2022. PMID: 35323034 Free PMC article. - Genetic and Functional Characterization of a Conjugative KpVP-2-Type Virulence Plasmid From a Clinical Klebsiella pneumoniae Strain.
Yang X, Liu X, Xu Y, Yang C, Chan EW, Shum HP, Chen S. Yang X, et al. Front Microbiol. 2022 Jul 22;13:914884. doi: 10.3389/fmicb.2022.914884. eCollection 2022. Front Microbiol. 2022. PMID: 35935210 Free PMC article. - Carbapenem Resistance-Encoding and Virulence-Encoding Conjugative Plasmids in Klebsiella pneumoniae.
Yang X, Dong N, Chan EW, Zhang R, Chen S. Yang X, et al. Trends Microbiol. 2021 Jan;29(1):65-83. doi: 10.1016/j.tim.2020.04.012. Epub 2020 May 21. Trends Microbiol. 2021. PMID: 32448764 Review. - Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms.
Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, Jeong BC, Lee SH. Lee CR, et al. Front Cell Infect Microbiol. 2017 Nov 21;7:483. doi: 10.3389/fcimb.2017.00483. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29209595 Free PMC article. Review.
Cited by
- Genetic diversity and evolution of the virulence plasmids encoding aerobactin and salmochelin in Klebsiella pneumoniae.
Tian D, Wang M, Zhou Y, Hu D, Ou HY, Jiang X. Tian D, et al. Virulence. 2021 Dec;12(1):1323-1333. doi: 10.1080/21505594.2021.1924019. Virulence. 2021. PMID: 33970792 Free PMC article. - Highly conserved composite transposon harbouring aerobactin iuc3 in Klebsiella pneumoniae from pigs.
Kaspersen H, Franklin-Alming FV, Hetland MAK, Bernhoff E, Löhr IH, Jiwakanon J, Urdahl AM, Leangapichart T, Sunde M. Kaspersen H, et al. Microb Genom. 2023 Feb;9(2):mgen000960. doi: 10.1099/mgen.0.000960. Microb Genom. 2023. PMID: 36820818 Free PMC article. - Large-scale genomic analysis of global Klebsiella pneumoniae plasmids reveals multiple simultaneous clusters of carbapenem-resistant hypervirulent strains.
Spadar A, Perdigão J, Campino S, Clark TG. Spadar A, et al. Genome Med. 2023 Jan 19;15(1):3. doi: 10.1186/s13073-023-01153-y. Genome Med. 2023. PMID: 36658655 Free PMC article. - Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen.
Gorrie CL, Mirčeta M, Wick RR, Judd LM, Lam MMC, Gomi R, Abbott IJ, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, Hunter PC, Pilcher DV, McGloughlin SA, Spelman DW, Wyres KL, Jenney AWJ, Holt KE. Gorrie CL, et al. Nat Commun. 2022 May 31;13(1):3017. doi: 10.1038/s41467-022-30717-6. Nat Commun. 2022. PMID: 35641522 Free PMC article. - Integration of phenotypic, qPCR and genome sequencing methodologies for the detection of antimicrobial resistance and virulence in clinical isolates of a tertiary hospital, India.
Vohra M, Babariya M, Parmar JS, Kamath N, Warghane A, Zala D. Vohra M, et al. 3 Biotech. 2023 Nov;13(11):368. doi: 10.1007/s13205-023-03797-4. Epub 2023 Oct 15. 3 Biotech. 2023. PMID: 37849769 Free PMC article.
References
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases