VFDB 2019: a comparative pathogenomic platform with an interactive web interface - PubMed (original) (raw)
VFDB 2019: a comparative pathogenomic platform with an interactive web interface
Bo Liu et al. Nucleic Acids Res. 2019.
Abstract
The virulence factor database (VFDB, http://www.mgc.ac.cn/VFs/) is devoted to providing the scientific community with a comprehensive warehouse and online platform for deciphering bacterial pathogenesis. The various combinations, organizations and expressions of virulence factors (VFs) are responsible for the diverse clinical symptoms of pathogen infections. Currently, whole-genome sequencing is widely used to decode potential novel or variant pathogens both in emergent outbreaks and in routine clinical practice. However, the efficient characterization of pathogenomic compositions remains a challenge for microbiologists or physicians with limited bioinformatics skills. Therefore, we introduced to VFDB an integrated and automatic pipeline, VFanalyzer, to systematically identify known/potential VFs in complete/draft bacterial genomes. VFanalyzer first constructs orthologous groups within the query genome and preanalyzed reference genomes from VFDB to avoid potential false positives due to paralogs. Then, it conducts iterative and exhaustive sequence similarity searches among the hierarchical prebuilt datasets of VFDB to accurately identify potential untypical/strain-specific VFs. Finally, via a context-based data refinement process for VFs encoded by gene clusters, VFanalyzer can achieve relatively high specificity and sensitivity without manual curation. In addition, a thoroughly optimized interactive web interface is introduced to present VFanalyzer reports in comparative pathogenomic style for easy online analysis.
Figures
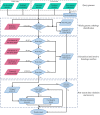
Figure 1.
The overall workflow of VFanalyzer data analysis processes. The green parallelograms are user-defined input data, and the red ones are curated datasets from the VFDB database.
Similar articles
- VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics.
Yang J, Chen L, Sun L, Yu J, Jin Q. Yang J, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D539-42. doi: 10.1093/nar/gkm951. Epub 2007 Nov 4. Nucleic Acids Res. 2008. PMID: 17984080 Free PMC article. - VFDB 2022: a general classification scheme for bacterial virulence factors.
Liu B, Zheng D, Zhou S, Chen L, Yang J. Liu B, et al. Nucleic Acids Res. 2022 Jan 7;50(D1):D912-D917. doi: 10.1093/nar/gkab1107. Nucleic Acids Res. 2022. PMID: 34850947 Free PMC article. - VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on.
Chen L, Zheng D, Liu B, Yang J, Jin Q. Chen L, et al. Nucleic Acids Res. 2016 Jan 4;44(D1):D694-7. doi: 10.1093/nar/gkv1239. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578559 Free PMC article. - Bacterial genomics and pathogen evolution.
Raskin DM, Seshadri R, Pukatzki SU, Mekalanos JJ. Raskin DM, et al. Cell. 2006 Feb 24;124(4):703-14. doi: 10.1016/j.cell.2006.02.002. Cell. 2006. PMID: 16497582 Review. - In silico Reconstruction of the Metabolic and Pathogenic Potential of Bacterial Genomes Using Subsystems.
McNeil LK, Aziz RK. McNeil LK, et al. Genome Dyn. 2009;6:21-34. doi: 10.1159/000235760. Epub 2009 Aug 19. Genome Dyn. 2009. PMID: 19696491 Review.
Cited by
- Characterization of Novel Bacteriophage vB_KpnP_ZX1 and Its Depolymerases with Therapeutic Potential for K57 Klebsiella pneumoniae Infection.
Li P, Ma W, Shen J, Zhou X. Li P, et al. Pharmaceutics. 2022 Sep 10;14(9):1916. doi: 10.3390/pharmaceutics14091916. Pharmaceutics. 2022. PMID: 36145665 Free PMC article. - The microbiome, resistome, and their co-evolution in sewage at a hospital for infectious diseases in Shanghai, China.
Ma Y, Wu N, Zhang T, Li Y, Cao L, Zhang P, Zhang Z, Zhu T, Zhang C. Ma Y, et al. Microbiol Spectr. 2024 Feb 6;12(2):e0390023. doi: 10.1128/spectrum.03900-23. Epub 2023 Dec 22. Microbiol Spectr. 2024. PMID: 38132570 Free PMC article. - Microbial Interdomain Interactions Delineate the Disruptive Intestinal Homeostasis in Clostridioides difficile Infection.
Herrera G, Arboleda JC, Pérez-Jaramillo JE, Patarroyo MA, Ramírez JD, Muñoz M. Herrera G, et al. Microbiol Spectr. 2022 Oct 26;10(5):e0050222. doi: 10.1128/spectrum.00502-22. Epub 2022 Sep 26. Microbiol Spectr. 2022. PMID: 36154277 Free PMC article. - Genetic diversity and variation in antimicrobial-resistance determinants of non-serotype 2 Streptococcus suis isolates from healthy pigs.
Kittiwan N, Calland JK, Mourkas E, Hitchings MD, Murray S, Tadee P, Tadee P, Duangsonk K, Meric G, Sheppard SK, Patchanee P, Pascoe B. Kittiwan N, et al. Microb Genom. 2022 Nov;8(11):mgen000882. doi: 10.1099/mgen.0.000882. Microb Genom. 2022. PMID: 36326658 Free PMC article. - Contraction and expansion dynamics: deciphering genomic underpinnings of growth rate and pathogenicity in Mycobacterium.
Zhu X, Lu Q, Li Y, Long Q, Zhang X, Long X, Cao D. Zhu X, et al. Front Microbiol. 2023 Nov 23;14:1292897. doi: 10.3389/fmicb.2023.1292897. eCollection 2023. Front Microbiol. 2023. PMID: 38075891 Free PMC article.
References
- Rohde H., Qin J., Cui Y., Li D., Loman N.J., Hentschke M., Chen W., Pu F., Peng Y., Li J. et al. . Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 2011; 365:718–724. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources