UniProt: a worldwide hub of protein knowledge - PubMed (original) (raw)
UniProt: a worldwide hub of protein knowledge
UniProt Consortium. Nucleic Acids Res. 2019.
Abstract
The UniProt Knowledgebase is a collection of sequences and annotations for over 120 million proteins across all branches of life. Detailed annotations extracted from the literature by expert curators have been collected for over half a million of these proteins. These annotations are supplemented by annotations provided by rule based automated systems, and those imported from other resources. In this article we describe significant updates that we have made over the last 2 years to the resource. We have greatly expanded the number of Reference Proteomes that we provide and in particular we have focussed on improving the number of viral Reference Proteomes. The UniProt website has been augmented with new data visualizations for the subcellular localization of proteins as well as their structure and interactions. UniProt resources are available under a CC-BY (4.0) license via the web at https://www.uniprot.org/.
Figures
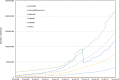
Figure 1.
Growth of UniProt sequences over the last decade.
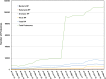
Figure 2.
Growth of the total number of Complete Proteomes and Reference Proteomes since 2015.
Figure 3.
Functional annotation describing human METTL14 (UniProtKB Q9HCE5).
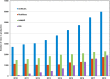
Figure 4.
Growth of curated automatic annotation rules within the UniRule system.
Figure 5.
Interaction matrix of the human Parkin protein.
Figure 6.
The subcellular localization view of a UniProt entry (UniProtKB P35670).
Figure 7.
The molecular structure of the Spike protein of the Human SARS coronavirus (PDB ID: 1WNC) structure as shown in the ProtVista protein viewer. The 3D viewer is interactively connected with the sequence level annotations in UniProt e.g. domains, PTMs and mutations. Note that the user can select from any of the structures that map to the protein entry.
Similar articles
- UniProt: the universal protein knowledgebase in 2021.
UniProt Consortium. UniProt Consortium. Nucleic Acids Res. 2021 Jan 8;49(D1):D480-D489. doi: 10.1093/nar/gkaa1100. Nucleic Acids Res. 2021. PMID: 33237286 Free PMC article. - UniProt: the Universal Protein Knowledgebase in 2023.
UniProt Consortium. UniProt Consortium. Nucleic Acids Res. 2023 Jan 6;51(D1):D523-D531. doi: 10.1093/nar/gkac1052. Nucleic Acids Res. 2023. PMID: 36408920 Free PMC article. - UniProt: the universal protein knowledgebase.
The UniProt Consortium. The UniProt Consortium. Nucleic Acids Res. 2017 Jan 4;45(D1):D158-D169. doi: 10.1093/nar/gkw1099. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899622 Free PMC article. - From protein sequences to 3D-structures and beyond: the example of the UniProt knowledgebase.
Hinz U; UniProt Consortium. Hinz U, et al. Cell Mol Life Sci. 2010 Apr;67(7):1049-64. doi: 10.1007/s00018-009-0229-6. Epub 2009 Dec 31. Cell Mol Life Sci. 2010. PMID: 20043185 Free PMC article. Review. - UniProt and Mass Spectrometry-Based Proteomics-A 2-Way Working Relationship.
Bowler-Barnett EH, Fan J, Luo J, Magrane M, Martin MJ, Orchard S; UniProt Consortium. Bowler-Barnett EH, et al. Mol Cell Proteomics. 2023 Aug;22(8):100591. doi: 10.1016/j.mcpro.2023.100591. Epub 2023 Jun 8. Mol Cell Proteomics. 2023. PMID: 37301379 Free PMC article. Review.
Cited by
- Protein A-like Peptide Design Based on Diffusion and ESM2 Models.
Zhao L, He Q, Song H, Zhou T, Luo A, Wen Z, Wang T, Lin X. Zhao L, et al. Molecules. 2024 Oct 21;29(20):4965. doi: 10.3390/molecules29204965. Molecules. 2024. PMID: 39459333 Free PMC article. - MCPdb: The bacterial microcompartment database.
Ochoa JM, Bair K, Holton T, Bobik TA, Yeates TO. Ochoa JM, et al. PLoS One. 2021 Mar 29;16(3):e0248269. doi: 10.1371/journal.pone.0248269. eCollection 2021. PLoS One. 2021. PMID: 33780471 Free PMC article. - Analysis of aqueous humor total antioxidant capacity and its correlation with corneal endothelial health.
Tsao YT, Wu WC, Chen KJ, Yeh LK, Hwang YS, Hsueh YJ, Chen HC, Cheng CM. Tsao YT, et al. Bioeng Transl Med. 2020 Dec 5;6(2):e10199. doi: 10.1002/btm2.10199. eCollection 2021 May. Bioeng Transl Med. 2020. PMID: 34027088 Free PMC article. - A multi-targeting drug design strategy for identifying potent anti-SARS-CoV-2 inhibitors.
Ren PX, Shang WJ, Yin WC, Ge H, Wang L, Zhang XL, Li BQ, Li HL, Xu YC, Xu EH, Jiang HL, Zhu LL, Zhang LK, Bai F. Ren PX, et al. Acta Pharmacol Sin. 2022 Feb;43(2):483-493. doi: 10.1038/s41401-021-00668-7. Epub 2021 Apr 27. Acta Pharmacol Sin. 2022. PMID: 33907306 Free PMC article. - Prediction of lncRNA-Protein Interactions via the Multiple Information Integration.
Chen Y, Fu X, Li Z, Peng L, Zhuo L. Chen Y, et al. Front Bioeng Biotechnol. 2021 Feb 25;9:647113. doi: 10.3389/fbioe.2021.647113. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 33718346 Free PMC article.
References
- Giraldo-Calderón G.I., Emrich S.J., MacCallum R.M., Maslen G., Dialynas E., Topalis P., Ho N., Gesing S. VectorBase Consortium . VectorBase Consortium. Madey G., et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015; 43:D707–D713. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM080646/GM/NIGMS NIH HHS/United States
- U41 HG002273/HG/NHGRI NIH HHS/United States
- UL1 TR001409/TR/NCATS NIH HHS/United States
- U01 GM120953/GM/NIGMS NIH HHS/United States
- P20 GM103446/GM/NIGMS NIH HHS/United States
- U41 HG007822/HG/NHGRI NIH HHS/United States
- BB/M011674/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- RG/13/5/30112/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources