Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein - PubMed (original) (raw)
Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein
Hannah Kleine-Weber et al. Sci Rep. 2018.
Abstract
The Middle East respiratory syndrome-related coronavirus (MERS-CoV) can cause severe disease and has pandemic potential. Therefore, development of antiviral strategies is an important task. The activation of the viral spike protein (S) by host cell proteases is essential for viral infectivity and the responsible enzymes are potential therapeutic targets. The cellular proteases furin, cathepsin L and TMPRSS2 can activate MERS-S and may cleave the S protein at two distinct sites, termed S1/S2 and S2'. Moreover, a potential cathepsin L cleavage site in MERS-S has been reported. However, the relative importance of these sites for MERS-S activation is incompletely understood. Here, we used mutagenic analysis and MERS-S-bearing vectors to study the contribution of specific cleavage sites to S protein-driven entry. We found that an intact S1/S2 site was only required for efficient entry into cells expressing endogenous TMPRSS2. In keeping with a previous study, pre-cleavage at the S1/S2 motif (RSVR) was important although not essential for subsequent MERS-S activation by TMPRSS2, and indirect evidence was obtained that this motif is processed by a protease depending on an intact RXXR motif, most likely furin. In contrast, the S2' site (RSAR) was required for robust viral entry into all cell lines tested and the integrity of one of the two arginines was sufficient for efficient entry. These findings suggest that cleavage at S2' is carried out by proteases recognizing a single arginine, most likely TMPRSS2 and cathepsin L. Finally, mutation of the proposed cathepsin L site did not impact viral entry and double mutation of S1/S2 and S2' site was compatible with cathepsin L- but not TMPRSS2-dependent host cell entry, indicating that cathepsin L can process the S protein at auxiliary sites. Collectively, our results indicate a rigid sequence requirement for S protein activation by TMPRSS2 but not cathepsin L.
Conflict of interest statement
The authors declare no competing interests.
Figures
Figure 1
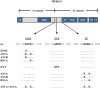
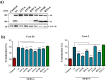
Domain organization and protease cleavage sites of MERS-S. MERS-S possesses two subunits, a surface subunit (S1) and a membrane-anchored subunit (S2). The S1 subunit harbors an N-terminal signal peptide (SP) and the receptor binding domain (RBD) while the S2 subunit contains domains required for membrane fusion, the fusion peptide (FP), two heptad repeats (HR1, HR2), and a transmembrane domain (TM). Moreover, the S2 subunit contains a cytoplasmic tail (CT). The S1/S2 cleavage site is located at the border between the S1 and S2 subunits while the S2′ site is located at the N-terminus of the FP. A proposed cleavage site for cathepsin L is located between the S1/S2 and S2′ sites (ECP, endosomal cysteine protease). The amino acid residues of the S1/S2, cathepsin L and S2′ sites are printed in bold and the mutations introduced into the cleavage sites are indicated.
Figure 2
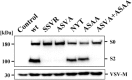
Incorporation of MERS-S proteins into rhabdoviral particles. Equal volumes of culture supernatants containing pseudoparticles harboring MERS-S wt or the indicated S protein mutants equipped with a C-terminal V5-tag were centrifuged and the pellets subjected to Western blot analysis, using an anti-V5 antibody. Arrow heads indicate bands corresponding to uncleaved precursor MERS-S (S0) and S2 subunit generated by cleavage at the S1/S2 border. The detection of VSV-M served as loading control. Shown is a representative Western blot from a total of twelve independent experiments
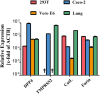
Figure 3
Expression of DPP4 and host cell proteases in target cell lines and lung tissue. Total cellular RNA was extracted from 293T, Caco-2 and Vero E6 cells, reverse-transcribed into cDNA and quantified for DPP4 and protease transcript numbers by quantitative RT-PCR. cDNA from human lung tissue was also included in the analysis. The numbers of DPP4/protease mRNA copies are shown relative to the housekeeping gene ß-actin (ACTB). Error bars indicate standard deviation (SD). Crosses indicate samples for which no transcripts were detected.
Figure 4
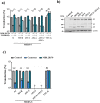
Requirement of the S1/S2 site for MERS-S-driven entry is cell type dependent, while the S2′ site is universally required. (a) Equal volumes of rhabdoviral vectors harboring MERS-S wt, the indicated S protein mutants, VSV-G or no glycoprotein at all (negative control) were used for transduction of Caco-2, Vero E6 cells or 293T cells that were either untreated or transfected with expression plasmid for DPP4. Transduction efficiency was quantified at 18 h post inoculation by measuring the activity of virus-encoded luciferase in cell lysates. Shown are the data from one representative experiment performed with quadruplicate samples, error bars indicate standard deviation (SD). Similar results were obtained in two separate experiments. (b) The combined results of three independent experiments carried out as described for panel a are shown. For normalization, transduction mediated by VSV particles harboring MERS-S wt was set as 100%. Error bars indicate standard error of the mean (SEM). Statistical significance of differences between transduction mediated by wt and mutant S proteins was analyzed using a paired, two-tailed students t-test (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns, not significant).
Figure 5
A single arginine at the S2′ site is sufficient for MERS-S activation. (a) Equal volumes of culture supernatants containing pseudoparticles harboring MERS-S wt or the indicated S protein mutants equipped with a C-terminal V5-tag were centrifuged and the pellets subjected to Western blot analysis, using an anti-V5 antibody. The results were confirmed in four separate experiments. (b) Vero E6 and Caco-2 cells were transduced with pseudoparticles harboring MERS-S wt, the indicated S protein mutants, VSV-G or no glycoprotein at all (negative control), and transduction efficiency was analyzed as described in the legend to Fig. 4. The average of nine separate experiments is shown, in which transduction mediated by MERS-S wt was set as 100%. Error bars indicate SEM. Statistical significance of differences between transduction mediated by wt and mutant S proteins was analyzed using a paired, two-tailed students t-test (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns, not significant).
Figure 6
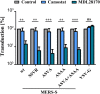
An intact S1/S2 site promotes but is not essential for MERS-S activation by TMPRSS2 (a) 293T cells were transfected with DPP4 plasmid or cotransfected with DPP4 and TMPRSS2 plasmid. At 24 h post transfection, cells were incubated with DMSO or cathepsin L inhibitor MDL28170 before being inoculated with pseudoparticles harboring MERS-S wt, the indicated S protein mutants or VSV-G. Transduction efficiency was quantified by measuring the activity of virus-encoded luciferase in cell lysates at 18 h post transduction. The average of three individual experiments is shown. Transduction of untreated, DPP4 transfected cells was set as 100%. Error bars indicate SEM. (b) 293T cells were cotransfected with TMPRSS2 plasmid and plasmids encoding MERS-S wt or the indicated S protein mutants equipped with a C-terminal V5-tag. Transfection of empty plasmid served as negative control. At 48 h post transfection, S protein expression in cell lysates was analyzed by Western blot. Bands representing uncleaved MERS-S (S0), the S2 subunit generated by cleavage at the S1/S2 site and an S2 fragment generated upon cleavage at the S2′ site are highlighted. Detection of ß-actin served as loading control. Similar results were obtained in two separate experiments. (c) Caco-2 cells were pre-incubated with the serine protease inhibitor camostat (bright blue) or the cathepsin L inhibitor MDL28170 (dark blue), or were control-treated with DMSO (gray). Subsequently, the cells were inoculated with equal volumes of preparations of pseudoparticles harboring MERS-S wt, the indicated S protein mutants or VSV-G. At 18 h post inoculation, transduction efficiency was quantified by measuring the activity of virus-encoded luciferase in cell lysates. The average of three independent experiments is shown. Transduction of control-treated cells was set as 100%. Crosses indicate samples for which no transduction above background levels was detected. Error bars indicate SEM. Statistical significance of differences between transduction of control-treated and inhibitor-treated cells was analyzed using a paired, two-tailed students t-test (**p ≤ 0.01; ***p ≤ 0.001; ns, not significant).
Figure 7
Cathepsin L activity is required for Vero E6 cell entry driven by a mutant S protein that lacks the S1/S2 and the S2′ sites. Vero E6 cells pre-incubated with serine protease inhibitor (camostat, bright blue), cathepsin L inhibitor (MDL28170, dark blue) or DMSO (control, gray) were inoculated with pseudoparticles as described for the Caco-2 cells in panel c of Fig. 6. At 18 h post inoculation, transduction efficiency was quantified by measuring the activity of virus-encoded luciferase in cell lysates. The average of three independent experiments is shown. Transduction of control-treated cells was set as 100%. Error bars indicate SEM. Statistical significance of differences between transduction of control-treated and inhibitor-treated cells was analyzed using a paired, two-tailed students t-test (**p ≤ 0.01; ***p ≤ 0.001; ns, not significant).
Similar articles
- Middle East Respiratory Syndrome Coronavirus Spike Protein Is Not Activated Directly by Cellular Furin during Viral Entry into Target Cells.
Matsuyama S, Shirato K, Kawase M, Terada Y, Kawachi K, Fukushi S, Kamitani W. Matsuyama S, et al. J Virol. 2018 Sep 12;92(19):e00683-18. doi: 10.1128/JVI.00683-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021905 Free PMC article. - Clinical Isolates of Human Coronavirus 229E Bypass the Endosome for Cell Entry.
Shirato K, Kanou K, Kawase M, Matsuyama S. Shirato K, et al. J Virol. 2016 Dec 16;91(1):e01387-16. doi: 10.1128/JVI.01387-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27733646 Free PMC article. - Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein.
Millet JK, Whittaker GR. Millet JK, et al. Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):15214-9. doi: 10.1073/pnas.1407087111. Epub 2014 Oct 6. Proc Natl Acad Sci U S A. 2014. PMID: 25288733 Free PMC article. - Proteolytic activation of SARS-CoV-2 spike protein.
Takeda M. Takeda M. Microbiol Immunol. 2022 Jan;66(1):15-23. doi: 10.1111/1348-0421.12945. Epub 2021 Oct 12. Microbiol Immunol. 2022. PMID: 34561887 Free PMC article. Review. - [Development of peptidic MERS-CoV entry inhibitors].
Xia S, Wang Q, Liu SW, Lu L, Jiang SB. Xia S, et al. Yao Xue Xue Bao. 2015 Dec;50(12):1513-9. Yao Xue Xue Bao. 2015. PMID: 27169270 Review. Chinese.
Cited by
- A cell-free platform to measure coronavirus membrane fusion.
Kicmal T, Qing E, Hawkins GM, Wilcox A, Gallagher T. Kicmal T, et al. STAR Protoc. 2023 Mar 6;4(2):102189. doi: 10.1016/j.xpro.2023.102189. Online ahead of print. STAR Protoc. 2023. PMID: 36952334 Free PMC article. - Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks.
Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA. Ashour HM, et al. Pathogens. 2020 Mar 4;9(3):186. doi: 10.3390/pathogens9030186. Pathogens. 2020. PMID: 32143502 Free PMC article. Review. - Enhancing immune protection against MERS-CoV: the synergistic effect of proteolytic cleavage sites and the fusion peptide and RBD domain targeting VLP immunization.
Oh J, Park U, Kim J, Jeon K, Kim C, Cho NH, Choi YS. Oh J, et al. Front Immunol. 2023 May 19;14:1201136. doi: 10.3389/fimmu.2023.1201136. eCollection 2023. Front Immunol. 2023. PMID: 37275866 Free PMC article. - The Transmembrane Protease TMPRSS2 as a Therapeutic Target for COVID-19 Treatment.
Wettstein L, Kirchhoff F, Münch J. Wettstein L, et al. Int J Mol Sci. 2022 Jan 25;23(3):1351. doi: 10.3390/ijms23031351. Int J Mol Sci. 2022. PMID: 35163273 Free PMC article. Review. - Spike proteins of novel MERS-coronavirus isolates from North- and West-African dromedary camels mediate robust viral entry into human target cells.
Kleine-Weber H, Pöhlmann S, Hoffmann M. Kleine-Weber H, et al. Virology. 2019 Sep;535:261-265. doi: 10.1016/j.virol.2019.07.016. Epub 2019 Jul 19. Virology. 2019. PMID: 31357164 Free PMC article.
References
- Lai, M. M. C., Perlman, S. & Anderson, L. J. In Fields virology Vol. 1 (eds D. M. Knipe & P. M. Howley) 1305–1336 (Lippincott, Wiliams & Wilkins, 2007).
- World Health, O. MERS situation update, March 2018 http://www.emro.who.int/pandemic-epidemic-diseases/mers-cov/mers-situati... (2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources