High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans) - PubMed (original) (raw)
High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans)
Weihua Liu et al. BMC Plant Biol. 2018.
Abstract
Background: Cold stress is one of the most severe abiotic stresses affecting the banana production. Although some miRNAs have been identified, little is known about the role of miRNAs in response to cold stress in banana, and up to date, there is no report about the role of miRNAs in the response to cold stress in the plants of the cultivated or wild bananas.
Result: Here, a cold-resistant line wild banana (Musa itinerans) from China was used to profile the cold-responsive miRNAs by RNA-seq during cold stress. Totally, 265 known mature miRNAs and 41 novel miRNAs were obtained. Cluster analysis of differentially expressed (DE) miRNAs indicated that some miRNAs were specific for chilling or 0 °C treated responses, and most of them were reported to be cold-responsive; however, some were seldom reported to be cold-responsive in response to cold stress, e.g., miR395, miR408, miR172, suggesting that they maybe play key roles in response to cold stress. The GO and KEGG pathway enrichment analysis of DE miRNAs targets indicated that there existed diversified cold-responsive pathways, and miR172 was found likely to play a central coordinating role in response to cold stress, especially in the regulation of CK2 and the circadian rhythm. Finally, qPCR assays indicated the related targets were negatively regulated by the tested DE miRNAs during cold stress in the wild banana.
Conclusions: In this study, the profiling of miRNAs by RNA-seq in response to cold stress in the plants of the wild banana (Musa itinerans) was reported for the first time. The results showed that there existed diversified cold-responsive pathways, which provided insight into the roles of miRNAs during cold stress, and would be helpful for alleviating cold stress and cold-resistant breeding in bananas.
Keywords: Cold stress; Musa itinerans; RNA-seq-based profiling; miR172; miRNA.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Fig. 1
Morphological changes of leaves from field plants and in vitro plants treated during cold stress
Fig. 2
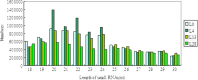
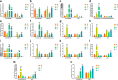
The length distribution of sequencing reads from 4 small RNA libraries in the wild banana. L0, L4, L13, L28: treated at 0 °C, 4 °C, 13 °C, 28 °C, respectively (the same below)
Fig. 3
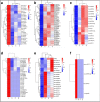
Heatmap of DE miRNAs during cold stress in the wild banana. The color represents miRNA expression values from 1.5 (the red corresponds to sRNAs with high expression) to − 1.5 (the blue corresponds to sRNAs with low expression). L0, L4, L13 and L28 correspond to the libraries obtained in the temperature 0 °C, 4 °C, 13 °C and 28 °C respectively. a up-regulation at 28 °C but down-regulation at other 3 temperatures; b: down-regulation at 28 °C but up-regulation at other 3 temperatures; c: down-regulation at 13 °C but up-regulation at other 3 temperatures; d: up-regulation at 4 °C but down-regulation at other 3 temperatures; e: down-regulation at 0 °C but up-regulation at other 3 temperatures; f: up-regulation at 0 °C but down-regulation at other 3 temperatures
Fig. 4
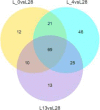
Venn diagrams of DE miRNAs among 3 groups during cold stress in the wild banana
Fig. 5
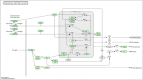
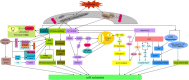
The plant circadian rhythm of the top 20 KEGG pathways in L_0 vs L28. The map was provided permission from Kanehisa laboratory [–180]. The nodes are marked in red background color indicating the DEGs is up-regulated expression, and the nodes are marked in green background color indicating the DEGs is down-regulated expression. On the other side, the white nodes indicated the genes are DEGs, but there is no distinguishment of the DEGs about the up- / down-regulated expression
Fig. 6
The qPCR validation of partial miRNAs and the target genes during cold stress in the wild banana. a-f. The expression patterns of the 6 miRNAs were opposite to those of the related targets. g-n. The expressions of miR172 and the related targets by qPCR (g-i: different sin3 gene members; j-n: _ada1_s). Duncan’s multiple range test: * P < 0.05; ** P < 0.01; the number of biological replicates = 3
Fig. 7
The diversified cold-responsive pathways of DE miRNAs targets during cold stress in the wild banana
Similar articles
- Genome-wide identification and characterization of mRNAs and lncRNAs involved in cold stress in the wild banana (Musa itinerans).
Liu W, Cheng C, Lin Y, XuHan X, Lai Z. Liu W, et al. PLoS One. 2018 Jul 9;13(7):e0200002. doi: 10.1371/journal.pone.0200002. eCollection 2018. PLoS One. 2018. PMID: 29985922 Free PMC article. - Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress.
Zhu H, Zhang Y, Tang R, Qu H, Duan X, Jiang Y. Zhu H, et al. BMC Genomics. 2019 Jan 10;20(1):33. doi: 10.1186/s12864-018-5395-1. BMC Genomics. 2019. PMID: 30630418 Free PMC article. - Genome-wide identification and characterization of the CKII gene family in the cultivated banana cultivar (Musa spp. cv Tianbaojiao) and the wild banana (Musa itinerans).
Liu W, Lin Z, Liu Y, Lin Y, Xu X, Lai Z. Liu W, et al. PLoS One. 2018 Jul 11;13(7):e0200149. doi: 10.1371/journal.pone.0200149. eCollection 2018. PLoS One. 2018. PMID: 29995937 Free PMC article. - Molecular cloning and expression analysis of KIN10 and cold-acclimation related genes in wild banana 'Huanxi' (Musa itinerans).
Liu W, Cheng C, Lai G, Lin Y, Lai Z. Liu W, et al. Springerplus. 2015 Dec 30;4:829. doi: 10.1186/s40064-015-1617-z. eCollection 2015. Springerplus. 2015. PMID: 26753116 Free PMC article. - Exploring diverse roles of micro RNAs in banana: Current status and future prospective.
Bhakta S, Tak H, Ganapathi TR. Bhakta S, et al. Physiol Plant. 2021 Dec;173(4):1323-1334. doi: 10.1111/ppl.13311. Epub 2020 Dec 22. Physiol Plant. 2021. PMID: 33305854 Review.
Cited by
- Combined analysis of mRNA and miRNA reveals the banana potassium absorption regulatory network and validation of miRNA160a.
Chen W, Dong T, Chen Y, Lin P, Wang C, Chen K, Tang Y, Wang M, Liu J, Yu H. Chen W, et al. Plant Mol Biol. 2022 Dec;110(6):531-543. doi: 10.1007/s11103-022-01304-6. Epub 2022 Aug 13. Plant Mol Biol. 2022. PMID: 35962899 - Epigenetic Mechanisms Driving Adaptation in Tropical and Subtropical Plants: Insights and Future Directions.
Miryeganeh M. Miryeganeh M. Plant Cell Environ. 2025 May;48(5):3487-3499. doi: 10.1111/pce.15370. Epub 2025 Jan 8. Plant Cell Environ. 2025. PMID: 39776407 Free PMC article. Review. - Transcriptome Analysis Revealed ZmPTOX1 Is Required for Seedling Development and Stress Tolerance in Maize.
Peng Y, Liang Z, Qing X, Wen M, Yuan Z, Chen Q, Du X, Gu R, Wang J, Li L. Peng Y, et al. Plants (Basel). 2024 Aug 23;13(17):2346. doi: 10.3390/plants13172346. Plants (Basel). 2024. PMID: 39273830 Free PMC article. - Characterization of the complete chloroplast genome of Sanming wild banana (Musa itinerans) and phylogenetic relationships.
Zhang YY, Liu F, Tian N, Che JR, Sun XL, Lai ZX, Cheng CZ. Zhang YY, et al. Mitochondrial DNA B Resour. 2019 Jul 17;4(2):2614-2616. doi: 10.1080/23802359.2019.1642167. Mitochondrial DNA B Resour. 2019. PMID: 33365650 Free PMC article. - Characterization of microRNAs and Target Genes in Musa acuminata subsp. burmannicoides, var. Calcutta 4 during Interaction with Pseudocercospora musae.
Rego ECS, Pinheiro TDM, Fonseca FCA, Gomes TG, Costa EC, Bastos LS, Alves GSC, Cotta MG, Amorim EP, Ferreira CF, Togawa RC, Costa MMDC, Grynberg P, Miller RNG. Rego ECS, et al. Plants (Basel). 2023 Mar 28;12(7):1473. doi: 10.3390/plants12071473. Plants (Basel). 2023. PMID: 37050099 Free PMC article.
References
- Lescot T. The genetic diversity of banana in figures. FruiTrop. 2011;189:58–62.
- Sabine A. FAO, editor. Food Outlook-Biannual Report on Global Food Markets. Rome: FAO; 2017. Opportunities and Challenges in the Banana Market; pp. 63–66.
- Lessard WO, Lessard WO. The Complete Book of Bananas. 1 1992.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources